Fast gapped-read alignment with Bowtie 2 (V体育官网入口)
- PMID: 22388286
- PMCID: PMC3322381
- DOI: 10.1038/nmeth.1923
Fast gapped-read alignment with Bowtie 2
"V体育平台登录" Abstract
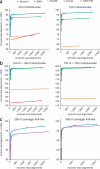
As the rate of sequencing increases, greater throughput is demanded from read aligners. The full-text minute index is often used to make alignment very fast and memory-efficient, but the approach is ill-suited to finding longer, gapped alignments VSports手机版. Bowtie 2 combines the strengths of the full-text minute index with the flexibility and speed of hardware-accelerated dynamic programming algorithms to achieve a combination of high speed, sensitivity and accuracy. .
Figures
References
-
- McKenna A, et al. Genome Res. 2010;20:1297–1303. - PMC (VSports最新版本) - PubMed
-
- Li H, Homer N. Brief. Bioinform. 2010;11:473–483. - "V体育安卓版" PMC - PubMed
-
- Ferragina P, Manzini G. Proc. 41st Annual Symposium on Foundations of Computer Science. IEEE Comput. Soc.; 2000. pp. 390–398.
Publication types
- Actions (VSports app下载)
- "V体育ios版" Actions
MeSH terms
- "VSports app下载" Actions
- VSports手机版 - Actions
- Actions (V体育ios版)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases (V体育官网入口)
Research Materials

