Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase
- PMID: 22203994
- PMCID: PMC3258621
- DOI: "VSports注册入口" 10.1073/pnas.1113251108
Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase
Abstract
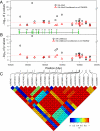
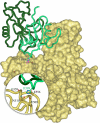
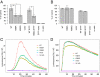
Systemic lupus erythematosus (SLE), the prototypic systemic autoimmune disease, is a debilitating multisystem autoimmune disorder characterized by chronic inflammation and extensive immune dysregulation in multiple organ systems, resulting in significant morbidity and mortality. Here, we present a multidisciplinary approach resulting in the identification of neutrophil cytosolic factor 2 (NCF2) as an important risk factor for SLE and the detailed characterization of its causal variant. We show that NCF2 is strongly associated with increased SLE risk in two independent populations: childhood-onset SLE and adult-onset SLE. The association between NCF2 and SLE can be attributed to a single nonsynonymous coding mutation in exon 12, the effect of which is the substitution of histidine-389 with glutamine (H389Q) in the PB1 domain of the NCF2 protein, with glutamine being the risk allele. Computational modeling suggests that the NCF2 H389Q mutation reduces the binding efficiency of NCF2 with the guanine nucleotide exchange factor Vav1. The model predicts that NCF2/H389 residue interacts with Vav1 residues E509, N510, E556, and G559 in the ZF domain of Vav1. Furthermore, replacing H389 with Q results in 1 VSports手机版. 5 kcal/mol weaker binding. To examine the effect of the NCF2 H389Q mutation on NADPH oxidase function, site-specific mutations at the 389 position in NCF2 were tested. Results show that an H389Q mutation causes a twofold decrease in reactive oxygen species production induced by the activation of the Vav-dependent Fcγ receptor-elicited NADPH oxidase activity. Our study completes the chain of evidence from genetic association to specific molecular function. .
Conflict of interest statement (V体育ios版)
The authors declare no conflict of interest.
Figures
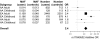



References
-
- Tan W, et al. BIOLUPUS Network. GENLES Network Association of PPP2CA polymorphisms with systemic lupus erythematosus susceptibility in multiple ethnic groups. Arthritis Rheum. 2011;63:2755–2763. - "VSports" PMC - PubMed
-
- Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–692. - VSports在线直播 - PMC - PubMed
-
- Harley JB, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. - PMC (V体育官网) - PubMed
-
- Jacob CO, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis Rheum. 2007;56:4164–4173. - PubMed
Publication types
"VSports在线直播" MeSH terms
- "VSports app下载" Actions
- "V体育2025版" Actions
- Actions (V体育ios版)
- Actions (V体育2025版)
- "V体育安卓版" Actions
- VSports手机版 - Actions
- "VSports手机版" Actions
- Actions (V体育官网入口)
- "V体育安卓版" Actions
- "VSports最新版本" Actions
Substances
- "VSports最新版本" Actions
- Actions (VSports在线直播)
- "VSports最新版本" Actions
Grants and funding
- AR049084/AR/NIAMS NIH HHS/United States
- R01 AI063274/AI/NIAID NIH HHS/United States
- P20 RR020143/RR/NCRR NIH HHS/United States
- R56 AI063274/AI/NIAID NIH HHS/United States
- P30 AR053483/AR/NIAMS NIH HHS/United States
- HL45635/HL/NHLBI NIH HHS/United States (VSports最新版本)
- AR042460/AR/NIAMS NIH HHS/United States
- AR043815/AR/NIAMS NIH HHS/United States (V体育平台登录)
- R01 AR057172/AR/NIAMS NIH HHS/United States
- R01 AR033062/AR/NIAMS NIH HHS/United States
- "V体育ios版" R01 AI024717/AI/NIAID NIH HHS/United States
- AR33062/AR/NIAMS NIH HHS/United States
- VSports - AI024717/AI/NIAID NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- AI08394/AI/NIAID NIH HHS/United States (VSports注册入口)
- RR020143/RR/NCRR NIH HHS/United States (VSports手机版)
- R01 HL045635/HL/NHLBI NIH HHS/United States
- N01 AR062277/AR/NIAMS NIH HHS/United States
- AI063274/AI/NIAID NIH HHS/United States
- "VSports app下载" R37 AI024717/AI/NIAID NIH HHS/United States
- R01 AR042460/AR/NIAMS NIH HHS/United States (VSports注册入口)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials (VSports最新版本)
Miscellaneous

