p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network
- PMID: 22184109
- PMCID: PMC3285336
- DOI: 10.1074/jbc.M111.328120 (VSports在线直播)
p63 regulates human keratinocyte proliferation via MYC-regulated gene network and differentiation commitment through cell adhesion-related gene network (V体育官网入口)
Abstract
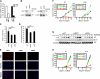
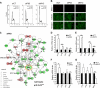
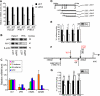
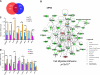
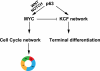
Although p63 and MYC are important in the control of epidermal homeostasis, the underlying molecular mechanisms governing keratinocyte proliferation or differentiation downstream of these two genes are not completely understood. By analyzing the transcriptional changes and phenotypic consequences of the loss of either p63 or MYC in human developmentally mature keratinocytes, we have characterized the networks acting downstream of these two genes to control epidermal homeostasis. We show that p63 is required to maintain growth and to commit to differentiation by two distinct mechanisms. Knockdown of p63 led to down-regulation of MYC via the Wnt/β-catenin and Notch signaling pathways and in turn reduced keratinocyte proliferation. We demonstrate that a p63-controlled keratinocyte cell fate network is essential to induce the onset of keratinocyte differentiation. This network contains several secreted proteins involved in cell migration/adhesion, including fibronectin 1 (FN1), interleukin-1β (IL1B), cysteine-rich protein 61 (CYR61), and jagged-1 (JAG1), that act downstream of p63 as key effectors to trigger differentiation. Our results characterized for the first time a connection between p63 and MYC and a cell adhesion-related network that controls differentiation VSports手机版. Furthermore, we show that the balance between the MYC-controlled cell cycle progression network and the p63-controlled cell adhesion-related network could dictate skin cell fate. .
Figures


References
-
- Fuchs E. (2007) Scratching the surface of skin development. Nature 445, 834–842 - V体育平台登录 - PMC - PubMed
-
- Fuchs E., Raghavan S. (2002) Getting under the skin of epidermal morphogenesis. Nat. Rev. Genet. 3, 199–209 - V体育ios版 - PubMed
-
- Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial, and epithelial development. Nature 398, 714–718 - PubMed (V体育ios版)
-
- Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 - PubMed
-
- Senoo M., Pinto F., Crum C. P., McKeon F. (2007) p63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536 - PubMed
"V体育官网" Publication types
MeSH terms
- VSports app下载 - Actions
- "V体育ios版" Actions
- V体育平台登录 - Actions
- V体育官网 - Actions
- V体育官网 - Actions
- V体育官网 - Actions
- "V体育官网入口" Actions
Substances
- Actions (V体育安卓版)
- VSports app下载 - Actions
- V体育官网入口 - Actions
- VSports注册入口 - Actions
"VSports在线直播" Associated data
- Actions (VSports注册入口)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

