Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity (VSports在线直播)
- PMID: 22144677
- PMCID: PMC3265867
- DOI: 10.1074/jbc.M111.305789
Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity
Abstract
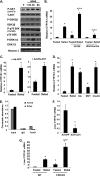
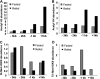
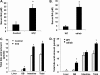
Bile acids facilitate postprandial absorption of nutrients. Bile acids also activate the farnesoid X receptor (FXR) and the G protein-coupled receptor TGR5 and play a major role in regulating lipid, glucose, and energy metabolism. Transgenic expression of cholesterol 7α-hydroxylase (CYP7A1) prevented high fat diet-induced diabetes and obesity in mice. In this study, we investigated the nutrient effects on bile acid synthesis. Refeeding of a chow diet to fasted mice increased CYP7A1 expression, bile acid pool size, and serum bile acids in wild type and humanized CYP7A1-transgenic mice. Chromatin immunoprecipitation assays showed that glucose increased histone acetylation and decreased histone methylation on the CYP7A1 gene promoter. Refeeding also induced CYP7A1 in fxr-deficient mice, indicating that FXR signaling did not play a role in postprandial regulation of bile acid synthesis. In streptozocin-induced type I diabetic mice and genetically obese type II diabetic ob/ob mice, hyperglycemia increased histone acetylation status on the CYP7A1 gene promoter, leading to elevated basal Cyp7a1 expression and an enlarged bile acid pool with altered bile acid composition. However, refeeding did not further increase CYP7A1 expression in diabetic mice. In summary, this study demonstrates that glucose and insulin are major postprandial factors that induce CYP7A1 gene expression and bile acid synthesis. Glucose induces CYP7A1 gene expression mainly by epigenetic mechanisms. In diabetic mice, CYP7A1 chromatin is hyperacetylated, and fasting to refeeding response is impaired and may exacerbate metabolic disorders in diabetes VSports手机版. .
Figures







References
-
- Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. (2009) Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89, 147–191 - PubMed (VSports注册入口)
-
- Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 - PubMed
-
- Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., Gerard R. D., Repa J. J., Mangelsdorf D. J., Kliewer S. A. (2005) Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 - PubMed
Publication types
- Actions (V体育2025版)
- Actions (VSports最新版本)
VSports最新版本 - MeSH terms
- "VSports最新版本" Actions
- VSports - Actions
- V体育安卓版 - Actions
- Actions (V体育安卓版)
- "VSports手机版" Actions
- VSports最新版本 - Actions
- VSports手机版 - Actions
- V体育安卓版 - Actions
- V体育官网入口 - Actions
- "V体育平台登录" Actions
- "VSports在线直播" Actions
Substances
- "VSports" Actions
- "VSports" Actions
- Actions (VSports注册入口)
- "V体育安卓版" Actions
VSports - Grants and funding
LinkOut - more resources
VSports手机版 - Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

