Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88
- PMID: 22115825
- PMCID: PMC3388726
- DOI: 10.1136/gutjnl-2011-301104
Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88
Abstract
Background: The gut microbiota has profound effects on host physiology but local host-microbial interactions in the gut are only poorly characterised and are likely to vary from the sparsely colonised duodenum to the densely colonised colon VSports手机版. Microorganisms are recognised by pattern recognition receptors such as Toll-like receptors, which signal through the adaptor molecule MyD88. .
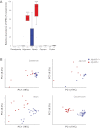
Methods: To identify host responses induced by gut microbiota along the length of the gut and whether these required MyD88, transcriptional profiles of duodenum, jejunum, ileum and colon were compared from germ-free and conventionally raised wild-type and Myd88-/- mice. The gut microbial ecology was assessed by 454-based pyrosequencing and viruses were analysed by PCR. V体育安卓版.
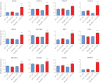
Results: The gut microbiota modulated the expression of a large set of genes in the small intestine and fewer genes in the colon but surprisingly few microbiota-regulated genes required MyD88 signalling. However, MyD88 was essential for microbiota-induced colonic expression of the antimicrobial genes Reg3β and Reg3γ in the epithelium, and Myd88 deficiency was associated with both a shift in bacterial diversity and a greater proportion of segmented filamentous bacteria in the small intestine. In addition, conventionally raised Myd88-/- mice had increased expression of antiviral genes in the colon, which correlated with norovirus infection in the colonic epithelium V体育ios版. .
Conclusion: This study provides a detailed description of tissue-specific host transcriptional responses to the normal gut microbiota along the length of the gut and demonstrates that the absence of MyD88 alters gut microbial ecology. VSports最新版本.
Conflict of interest statement
Figures

VSports在线直播 - Comment in
-
Regulation of host gene expression by gut microbiota.Gastroenterology. 2013 Apr;144(4):841-4. doi: 10.1053/j.gastro.2013.02.028. Epub 2013 Feb 24. Gastroenterology. 2013. PMID: 23462129 No abstract available.
References
-
- Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65 - VSports - PMC - PubMed
-
- Wostmann BS. The germfree animal in nutritional studies. Annu Rev Nutr 1981;1:257–79 - PubMed
"V体育安卓版" Publication types
- "V体育ios版" Actions
MeSH terms
- "V体育安卓版" Actions
- Actions (VSports最新版本)
- "VSports在线直播" Actions
- "V体育官网入口" Actions
- VSports app下载 - Actions
- V体育ios版 - Actions
- "VSports app下载" Actions
- VSports在线直播 - Actions
- Actions (V体育ios版)
Substances
- "V体育官网" Actions
VSports app下载 - Associated data
- Actions
VSports手机版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育安卓版 - Molecular Biology Databases
