From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay
- PMID: 22022604
- PMCID: PMC3195687
- DOI: 10.1371/journal.pone.0026332
From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay (V体育安卓版)
VSports手机版 - Abstract
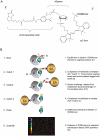
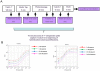
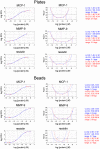
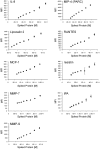
Recently, we reported a SOMAmer-based, highly multiplexed assay for the purpose of biomarker identification. To enable seamless transition from highly multiplexed biomarker discovery assays to a format suitable and convenient for diagnostic and life-science applications, we developed a streamlined, plate-based version of the assay. The plate-based version of the assay is robust, sensitive (sub-picomolar), rapid, can be highly multiplexed (upwards of 60 analytes), and fully automated. We demonstrate that quantification by microarray-based hybridization, Luminex bead-based methods, and qPCR are each compatible with our platform, further expanding the breadth of proteomic applications for a wide user community. VSports手机版.
Conflict of interest statement
Competing Interests: The authors have read the journal's policy and have the following conflict: the authors are all employees of SomaLogic, Inc. This fact does not alter the authors′ adherence to PLoS One policies on sharing of data and materials V体育安卓版.
Figures
References
-
- O'Farrell PH. The pre-omics era: the early days of two-dimensional gels. Proteomics. 2008;8:4842–4852. - PMC (V体育平台登录) - PubMed
-
- Zichi D, Eaton B, Singer B, Gold L. Proteomics and diagnostics: Let's Get Specific, again. Current Opinion in Chemical Biology. 2008;12:78–85. - PubMed (V体育官网)
-
- Ellington D, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818. - PubMed
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505. - PubMed
"V体育官网入口" Publication types
MeSH terms
- V体育官网 - Actions
- Actions (V体育官网入口)
- Actions (V体育平台登录)
- "VSports注册入口" Actions
- "V体育官网" Actions
- V体育2025版 - Actions
- Actions (V体育安卓版)
- "V体育官网入口" Actions
- "VSports手机版" Actions
- Actions (V体育ios版)
Substances
- V体育官网 - Actions
- Actions (V体育ios版)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

