Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine
- PMID: 22018469
- PMCID: "VSports app下载" PMC3225703
- DOI: 10.1016/j.immuni.2011.08.013 (V体育平台登录)
Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine
Abstract
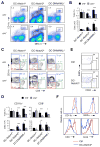
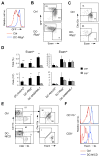
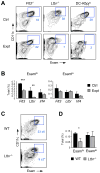
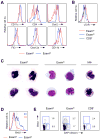
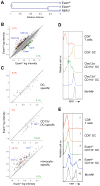
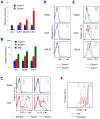
Dendritic cells (DCs) in tissues and lymphoid organs comprise distinct functional subsets that differentiate in situ from circulating progenitors VSports手机版. Tissue-specific signals that regulate DC subset differentiation are poorly understood. We report that DC-specific deletion of the Notch2 receptor caused a reduction of DC populations in the spleen. Within the splenic CD11b(+) DC subset, Notch signaling blockade ablated a distinct population marked by high expression of the adhesion molecule Esam. The Notch-dependent Esam(hi) DC subset required lymphotoxin beta receptor signaling, proliferated in situ, and facilitated CD4(+) T cell priming. The Notch-independent Esam(lo) DCs expressed monocyte-related genes and showed superior cytokine responses. In addition, Notch2 deletion led to the loss of CD11b(+)CD103(+) DCs in the intestinal lamina propria and to a corresponding decrease of IL-17-producing CD4(+) T cells in the intestine. Thus, Notch2 is a common differentiation signal for T cell-priming CD11b(+) DC subsets in the spleen and intestine. .
Copyright © 2011 Elsevier Inc V体育安卓版. All rights reserved. .
"V体育2025版" Figures

References
-
- Bar-On L, Birnberg T, Lewis KL, Edelson BT, Bruder D, Hildner K, Buer J, Murphy KM, Reizis B, Jung S. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:14745–14750. - PMC (V体育官网) - PubMed
-
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. - VSports注册入口 - PubMed
-
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. - VSports app下载 - PubMed
Publication types (V体育平台登录)
- VSports - Actions
MeSH terms
- Actions (VSports在线直播)
- "VSports手机版" Actions
- "V体育官网入口" Actions
- VSports - Actions
- Actions (VSports最新版本)
- V体育官网 - Actions
- V体育官网 - Actions
- V体育2025版 - Actions
- VSports最新版本 - Actions
- "VSports最新版本" Actions
- "V体育安卓版" Actions
- VSports在线直播 - Actions
"V体育平台登录" Substances
- V体育ios版 - Actions
- VSports在线直播 - Actions
- VSports在线直播 - Actions
- "V体育官网入口" Actions
Associated data (V体育官网)
- "VSports手机版" Actions
VSports注册入口 - Grants and funding
- R37 AI072571/AI/NIAID NIH HHS/United States
- R00 DK085329/DK/NIDDK NIH HHS/United States
- DK085329/DK/NIDDK NIH HHS/United States (V体育官网入口)
- AI007161/AI/NIAID NIH HHS/United States
- T32 HD055165/HD/NICHD NIH HHS/United States
- VSports手机版 - T32 AI007161/AI/NIAID NIH HHS/United States
- R01 AI072571/AI/NIAID NIH HHS/United States
- R21 AI067804/AI/NIAID NIH HHS/United States
- MOP 67157/CAPMC/ CIHR/Canada
- K99 DK085329/DK/NIDDK NIH HHS/United States (VSports在线直播)
- AI072571/AI/NIAID NIH HHS/United States
- HD055165/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases (VSports在线直播)
Research Materials (VSports在线直播)
Miscellaneous

