Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice
- PMID: 22006328
- PMCID: PMC3219119 (V体育官网)
- DOI: V体育平台登录 - 10.1073/pnas.1110905108
Iron deficiency drives an autosomal dominant hypophosphatemic rickets (ADHR) phenotype in fibroblast growth factor-23 (Fgf23) knock-in mice
Abstract
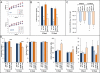
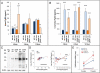
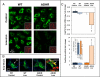
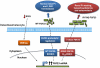
Autosomal dominant hypophosphatemic rickets (ADHR) is unique among the disorders involving Fibroblast growth factor 23 (FGF23) because individuals with R176Q/W and R179Q/W mutations in the FGF23 (176)RXXR(179)/S(180) proteolytic cleavage motif can cycle from unaffected status to delayed onset of disease. This onset may occur in physiological states associated with iron deficiency, including puberty and pregnancy. To test the role of iron status in development of the ADHR phenotype, WT and R176Q-Fgf23 knock-in (ADHR) mice were placed on control or low-iron diets. Both the WT and ADHR mice receiving low-iron diet had significantly elevated bone Fgf23 mRNA. WT mice on a low-iron diet maintained normal serum intact Fgf23 and phosphate metabolism, with elevated serum C-terminal Fgf23 fragments. In contrast, the ADHR mice on the low-iron diet had elevated intact and C-terminal Fgf23 with hypophosphatemic osteomalacia. We used in vitro iron chelation to isolate the effects of iron deficiency on Fgf23 expression VSports手机版. We found that iron chelation in vitro resulted in a significant increase in Fgf23 mRNA that was dependent upon Mapk. Thus, unlike other syndromes of elevated FGF23, our findings support the concept that late-onset ADHR is the product of gene-environment interactions whereby the combined presence of an Fgf23-stabilizing mutation and iron deficiency can lead to ADHR. .
Conflict of interest statement
Conflict of interest statement: K. E V体育安卓版. W. receives royalties for licensing the FGF23 gene to Kyowa Hakko Kirin Co. Ltd. , and M. P. is involved in a clinical trial with Kyowa Hakko Kirin Co. Ltd.
"V体育平台登录" Figures


References
-
- Econs MJ, McEnery PT. Autosomal dominant hypophosphatemic rickets/osteomalacia: Clinical characterization of a novel renal phosphate-wasting disorder. J Clin Endocrinol Metab. 1997;82:674–681. - PubMed
-
- ADHR Consortium Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat Genet. 2000;26:345–348. - PubMed (VSports)
-
- White KE, et al. Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney Int. 2001;60:2079–2086. - PubMed
-
- Shimada T, et al. Mutant FGF-23 responsible for autosomal dominant hypophosphatemic rickets is resistant to proteolytic cleavage and causes hypophosphatemia in vivo. Endocrinology. 2002;143:3179–3182. - PubMed (V体育平台登录)
-
- Urakawa I, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–774. - PubMed
Publication types
- V体育安卓版 - Actions
MeSH terms
- Actions (VSports手机版)
- VSports - Actions
- VSports手机版 - Actions
- Actions (VSports手机版)
- "V体育官网入口" Actions
- "VSports手机版" Actions
- V体育2025版 - Actions
- "VSports app下载" Actions
- V体育安卓版 - Actions
- Actions (V体育平台登录)
Substances
- "VSports手机版" Actions
- Actions (V体育安卓版)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical (V体育安卓版)
Molecular Biology Databases

