V体育安卓版 - Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types
- PMID: 21867929
- PMCID: V体育ios版 - PMC3169184
- DOI: "V体育2025版" 10.1016/j.immuni.2011.08.007
Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types
Abstract
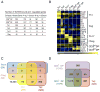
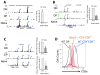
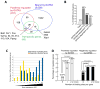
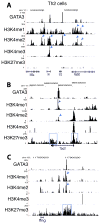
The transcription factor GATA3 plays an essential role during T cell development and T helper 2 (Th2) cell differentiation VSports手机版. To understand GATA3-mediated gene regulation, we identified genome-wide GATA3 binding sites in ten well-defined developmental and effector T lymphocyte lineages. In the thymus, GATA3 directly regulated many critical factors, including Th-POK, Notch1, and T cell receptor subunits. In the periphery, GATA3 induced a large number of Th2 cell-specific as well as Th2 cell-nonspecific genes, including several transcription factors. Our data also indicate that GATA3 regulates both active and repressive histone modifications of many target genes at their regulatory elements near GATA3 binding sites. Overall, although GATA3 binding exhibited both shared and cell-specific patterns among various T cell lineages, many genes were either positively or negatively regulated by GATA3 in a cell type-specific manner, suggesting that GATA3-mediated gene regulation depends strongly on cofactors existing in different T cells. .
Copyright © 2011 Elsevier Inc V体育安卓版. All rights reserved. .
Figures



References
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. - V体育ios版 - PMC - PubMed
Publication types
- "VSports注册入口" Actions
MeSH terms
- "V体育平台登录" Actions
- Actions (V体育ios版)
- "VSports手机版" Actions
- Actions (VSports)
- "VSports手机版" Actions
- "V体育官网" Actions
- "VSports手机版" Actions
- "V体育安卓版" Actions
- "VSports手机版" Actions
- "V体育官网" Actions
- "VSports手机版" Actions
- "V体育ios版" Actions
- Actions (VSports在线直播)
- "V体育官网入口" Actions
- "V体育官网" Actions
- Actions (VSports最新版本)
- "VSports最新版本" Actions
Substances
- V体育安卓版 - Actions
- V体育官网 - Actions
- VSports在线直播 - Actions
VSports注册入口 - Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

