"VSports app下载" Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume
- PMID: 21814288
- PMCID: PMC3260509
- DOI: "V体育ios版" 10.1038/ismej.2011.91
"V体育官网入口" Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume
V体育ios版 - Abstract
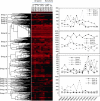
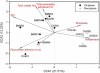
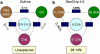
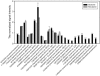
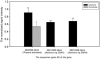
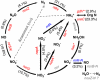
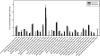
The Deepwater Horizon oil spill in the Gulf of Mexico is the deepest and largest offshore spill in the United State history and its impacts on marine ecosystems are largely unknown. Here, we showed that the microbial community functional composition and structure were dramatically altered in a deep-sea oil plume resulting from the spill VSports手机版. A variety of metabolic genes involved in both aerobic and anaerobic hydrocarbon degradation were highly enriched in the plume compared with outside the plume, indicating a great potential for intrinsic bioremediation or natural attenuation in the deep sea. Various other microbial functional genes that are relevant to carbon, nitrogen, phosphorus, sulfur and iron cycling, metal resistance and bacteriophage replication were also enriched in the plume. Together, these results suggest that the indigenous marine microbial communities could have a significant role in biodegradation of oil spills in deep-sea environments. .
Figures
References
-
- Aitken CM, Jones DM, Larter SR. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature. 2004;431:291–294. - PubMed
-
- Anderson M. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
-
- Barbeau K, Rue EL, Bruland KW, Butler A. Photochemical cycling of iron in the surface ocean mediated by microbial iron(III)-binding ligands. Nature. 2001a;413:409–413. - PubMed
-
- Barbeau K, Zhang G, Live DH, Butler A. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J Am Chem Soc. 2001b;124:378–379. - PubMed
Publication types
- VSports注册入口 - Actions
V体育安卓版 - MeSH terms
- Actions (VSports在线直播)
- Actions (VSports)
- "V体育2025版" Actions
- Actions (V体育官网)
- Actions (V体育ios版)
Substances
- V体育官网 - Actions
VSports手机版 - LinkOut - more resources
VSports app下载 - Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

