De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities (VSports在线直播)
- PMID: 21716304
- PMCID: PMC3246234
- DOI: 10.1038/ismej.2011.78
De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities
Abstract
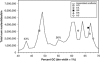
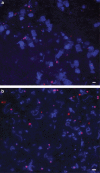
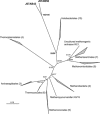
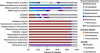
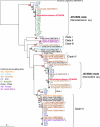
This study describes reconstruction of two highly unusual archaeal genomes by de novo metagenomic assembly of multiple, deeply sequenced libraries from surface waters of Lake Tyrrell (LT), a hypersaline lake in NW Victoria, Australia. Lineage-specific probes were designed using the assembled genomes to visualize these novel archaea, which were highly abundant in the 0. 1-0. 8 μm size fraction of lake water samples. Gene content and inferred metabolic capabilities were highly dissimilar to all previously identified hypersaline microbial species VSports手机版. Distinctive characteristics included unique amino acid composition, absence of Gvp gas vesicle proteins, atypical archaeal metabolic pathways and unusually small cell size (approximately 0. 6 μm diameter). Multi-locus phylogenetic analyses demonstrated that these organisms belong to a new major euryarchaeal lineage, distantly related to halophilic archaea of class Halobacteria. Consistent with these findings, we propose creation of a new archaeal class, provisionally named 'Nanohaloarchaea'. In addition to their high abundance in LT surface waters, we report the prevalence of Nanohaloarchaea in other hypersaline environments worldwide. The simultaneous discovery and genome sequencing of a novel yet ubiquitous lineage of uncultivated microorganisms demonstrates that even historically well-characterized environments can reveal unexpected diversity when analyzed by metagenomics, and advances our understanding of the ecology of hypersaline environments and the evolutionary history of the archaea. .
Figures

References
-
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. - PubMed
-
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, et al. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006;314:1933–1935. - PubMed
Publication types
MeSH terms
- Actions (VSports在线直播)
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- "V体育官网" Actions
- "VSports在线直播" Actions
- Actions (VSports手机版)
Associated data
- "VSports在线直播" Actions
- Actions
- "V体育ios版" Actions
- Actions
- "VSports手机版" Actions
- "VSports注册入口" Actions
- Actions (VSports最新版本)
- Actions
- Actions
- Actions (V体育官网)
- V体育官网 - Actions
- Actions
- V体育官网入口 - Actions
- Actions
- Actions
- Actions
- "VSports在线直播" Actions
- Actions
- "VSports手机版" Actions
- "V体育安卓版" Actions
- "VSports在线直播" Actions
- V体育平台登录 - Actions
- VSports手机版 - Actions
- VSports app下载 - Actions
- V体育安卓版 - Actions
- Actions
- V体育ios版 - Actions
- "V体育官网" Actions
- "V体育平台登录" Actions
- Actions
- Actions (VSports app下载)
"VSports app下载" Grants and funding
V体育官网 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

