"V体育安卓版" Chromatin insulator elements block transgene silencing in engineered human embryonic stem cell lines at a defined chromosome 13 locus
- PMID: 21699412
- PMCID: PMC3258440
- DOI: 10.1089/scd.2011.0163
Chromatin insulator elements block transgene silencing in engineered human embryonic stem cell lines at a defined chromosome 13 locus
V体育官网 - Abstract
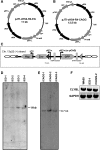
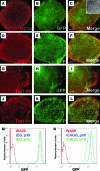
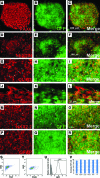
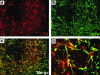
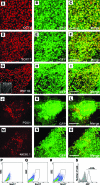
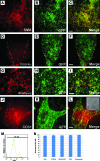
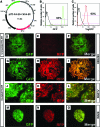
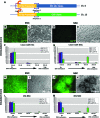
Lineage reporters of human embryonic stem cell (hESC) lines are useful for differentiation studies and drug screening. Previously, we created reporter lines driven by an elongation factor 1 alpha (EF1α) promoter at a chromosome 13q32. 3 locus in the hESC line WA09 and an abnormal hESC line BG01V in a site-specific manner. Expression of reporters in these lines was maintained in long-term culture at undifferentiated state. However, when these cells were differentiated into specific lineages, reduction in reporter expression was observed, indicating transgene silencing. To develop an efficient and reliable genetic engineering strategy in hESCs, we used chromatin insulator elements to flank single-copy transgenes and integrated the combined expression constructs via PhiC31/R4 integrase-mediated recombination technology to the chromosome 13 locus precisely. Two copies of cHS4 double-insulator sequences were placed adjacent to both 5' and 3' of the promoter reporter constructs. The green fluorescent protein (GFP) gene was driven by EF1α or CMV early enhancer/chicken β actin (CAG) promoter. In the engineered hESC lines, for both insulated CAG-GFP and EF1α-GFP, constitutive expression at the chromosome 13 locus was maintained during prolonged culture and in directed differentiation assays toward diverse types of neurons, pancreatic endoderm, and mesodermal progeny. In particular, described here is the first normal hESC fluorescent reporter line that robustly expresses GFP in both the undifferentiated state and throughout dopaminergic lineage differentiation. The dual strategy of utilizing insulator sequences and integration at the constitutive chromosome 13 locus ensures appropriate transgene expression VSports手机版. This is a valuable tool for lineage development study, gain- and loss-of-function experiments, and human disease modeling using hESCs. .
Figures
References
-
- Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
-
- Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. - PubMed
-
- Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. - PubMed
-
- Irion S. Luche H. Gadue P. Fehling HJ. Kennedy M. Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477–1482. - PubMed
-
- Costa M. Dottori M. Ng E. Hawes SM. Sourris K. Jamshidi P. Pera MF. Elefanty AG. Stanley EG. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat Methods. 2005;2:259–260. - PubMed
Publication types
- V体育官网 - Actions
- "VSports" Actions
MeSH terms
- VSports手机版 - Actions
- "V体育平台登录" Actions
- "V体育2025版" Actions
- VSports手机版 - Actions
- "V体育官网" Actions
- Actions (V体育2025版)
- "V体育2025版" Actions
- Actions (V体育安卓版)
- "V体育官网入口" Actions
- V体育安卓版 - Actions
Substances
- V体育安卓版 - Actions
- "VSports最新版本" Actions
- "V体育安卓版" Actions
"VSports" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
"V体育官网入口" Research Materials

