ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
- PMID: 21666679
- PMCID: PMC3130887
- DOI: 10.1038/nsmb.2062
ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
"V体育官网入口" Abstract
ATR-X (alpha-thalassemia/mental retardation, X-linked) syndrome is a human congenital disorder that causes severe intellectual disabilities. Mutations in the ATRX gene, which encodes an ATP-dependent chromatin-remodeler, are responsible for the syndrome. Approximately 50% of the missense mutations in affected persons are clustered in a cysteine-rich domain termed ADD (ATRX-DNMT3-DNMT3L, ADD(ATRX)), whose function has remained elusive. Here we identify ADD(ATRX) as a previously unknown histone H3-binding module, whose binding is promoted by lysine 9 trimethylation (H3K9me3) but inhibited by lysine 4 trimethylation (H3K4me3) VSports手机版. The cocrystal structure of ADD(ATRX) bound to H3(1-15)K9me3 peptide reveals an atypical composite H3K9me3-binding pocket, which is distinct from the conventional trimethyllysine-binding aromatic cage. Notably, H3K9me3-pocket mutants and ATR-X syndrome mutants are defective in both H3K9me3 binding and localization at pericentromeric heterochromatin; thus, we have discovered a unique histone-recognition mechanism underlying the ATR-X etiology. .
Conflict of interest statement
Y. S is a co-founder of Constellation Pharmaceuticals. D. J. Patel is on the Epigenetics Advisory Board of Epinova-Glaxo V体育安卓版. The remaining authors declare no competing financial interests.
Figures
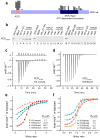
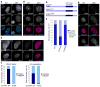

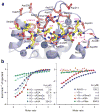


References
-
- Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. - PubMed
-
- Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. - VSports在线直播 - PMC - PubMed
-
- Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. - PubMed
Publication types
- VSports app下载 - Actions
MeSH terms
- Actions (VSports注册入口)
- "V体育官网" Actions
- Actions (VSports最新版本)
- "V体育官网" Actions
- "VSports手机版" Actions
- V体育安卓版 - Actions
Substances
- VSports最新版本 - Actions
- V体育平台登录 - Actions
- Actions (V体育官网入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

