"VSports最新版本" Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data
- PMID: 21633356
- PMCID: "VSports" PMC3400429
- DOI: "V体育安卓版" 10.1038/nbt.1873
Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data
Abstract
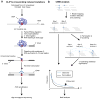
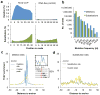
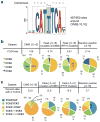
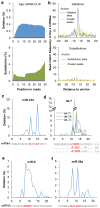
Mammalian RNA complexity is regulated through interactions of RNA-binding proteins (RBPs) with their target transcripts. High-throughput sequencing together with UV-crosslinking and immunoprecipitation (HITS-CLIP) is able to globally map RBP-binding footprint regions at a resolution of ~30-60 nucleotides. Here we describe a systematic way to analyze HITS-CLIP data to identify exact crosslink sites, and thereby determine protein-RNA interactions at single-nucleotide resolution. We found that reverse transcriptase used in CLIP frequently skips the crosslinked amino-acid-RNA adduct, resulting in a nucleotide deletion. Genome-wide analysis of these crosslinking-induced mutation sites (CIMS) in HITS-CLIP data for Nova and Argonaute (Ago) proteins in mouse brain tissue revealed deletions in ~8-20% of mRNA tags, which mapped to Nova and Ago binding sites on mRNA or miRNA. CIMS analysis provides a general and more precise means of mapping protein-RNA interactions than currently available methods and insight into the biochemical properties of such interactions in living tissues. VSports手机版.
Figures

References
-
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. - PMC (VSports在线直播) - PubMed
-
- Dredge BK, Darnell RB. Nova regulates GABAA receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol. 2003;23:4687–4700. - VSports - PMC - PubMed
-
- Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. - V体育平台登录 - PMC - PubMed
Publication types
- "V体育2025版" Actions
MeSH terms
- "VSports注册入口" Actions
- "V体育安卓版" Actions
- VSports app下载 - Actions
- VSports手机版 - Actions
- "V体育官网" Actions
- "V体育2025版" Actions
- Actions (VSports)
- Actions (V体育平台登录)
- Actions (VSports手机版)
- Actions (VSports手机版)
- V体育官网入口 - Actions
"V体育安卓版" Substances
- Actions (VSports注册入口)
- Actions (VSports注册入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

