Nrf2-dependent induction of NQO1 in mouse aortic endothelial cells overexpressing catalase (VSports注册入口)
- PMID: 21569840
- PMCID: PMC3109219
- DOI: 10.1016/j.freeradbiomed.2011.04.020
Nrf2-dependent induction of NQO1 in mouse aortic endothelial cells overexpressing catalase (VSports最新版本)
Abstract (V体育2025版)
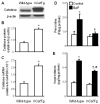
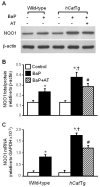
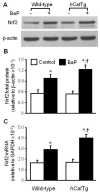
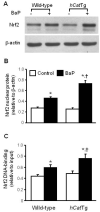
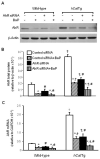
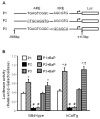
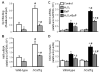
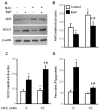
Overexpression of catalase has been shown to accelerate benzo(a)pyrene (BaP) detoxification in mouse aortic endothelial cells (MAECs). NAD(P)H:quinone oxidoreductase-1 (NQO1) is an enzyme that catalyzes BaP-quinone detoxification. Aryl hydrocarbon receptor (AhR) and nuclear factor erythroid 2-related factor-2 (Nrf2) are transcription factors that control NQO1 expression. Here, we investigated the effects of catalase overexpression on NQO1, Nrf2, and AhR expression. The levels of NQO1 mRNA and protein were comparable in MAECs isolated from wild-type and transgenic mice that overexpress human catalase (hCatTg). BaP treatment increased NQO1 mRNA and protein levels in both groups, with a significantly greater induction in hCatTg MAECs than in wild-type cells. BaP-induced NQO1 promoter activity was dramatically higher in hCatTg MAECs than in wild-type cells. Our data also showed that the basal level of AhR and the BaP-induced level of Nrf2 were significantly higher in hCatTg MAECs than in wild-type cells VSports手机版. Inhibition of specificity protein-1 (Sp1) binding to the AhR promoter region by mithramycin A reversed the enhancing effect of catalase overexpression on AhR expression. Knockdown of AhR by RNA interference diminished BaP-induced expression of Nrf2 and NQO1. Knockdown of Nrf2 significantly decreased NQO1 mRNA and protein levels in cells with or without BaP treatment. NQO1 promoter activity was abrogated by mutation of the Nrf2-binding site in this promoter. In contrast, mutation of the AhR-binding site in the NQO1 promoter did not affect the promoter activity. These results suggest that catalase overexpression upregulates BaP-induced NQO1 expression by enhancing the Sp1-AhR-Nrf2 signaling cascade. .
Copyright © 2011 Elsevier Inc V体育安卓版. All rights reserved. .
Figures





References
-
- Lind C. Formation of benzo[a]pyrene-3,6-quinol mono- and diglucuronides in rat liver microsomes. Arch. Biochem. Biophys. 1985;240:226–235. - "V体育安卓版" PubMed
-
- Joseph P, Jaiswal AK. NAD(P)H:quinone oxidoreductase1 (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8413–8417. - V体育官网 - PMC - PubMed
-
- Kann S, Huang MY, Estes C, Reichard JF, Sartor MA, Xia Y, Puga A. Arsenite-induced aryl hydrocarbon receptor nuclear translocation results in additive induction of phase I genes and synergistic induction of phase II genes. Mol. Pharmacol. 2005;68:336–346. - PubMed
-
- Ma Q, Kinneer K, Bi Y, Chan JY, Kan YW. Induction of murine NAD(P)H:quinone oxidoreductase by 2,3,7,8-tetrachlorodibenzo-p-dioxin requires the CNC (cap ‘n’ collar) basic leucine zipper transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2): cross-interaction between AhR (aryl hydrocarbon receptor) and Nrf2 signal transduction. Biochem. J. 2004;377:205–213. - PMC - PubMed
-
- Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm. Res. 2006;23:2586–2594. - PubMed
Publication types
- Actions (V体育官网入口)
MeSH terms
- VSports手机版 - Actions
- Actions (VSports app下载)
- V体育官网 - Actions
- V体育平台登录 - Actions
- Actions (VSports手机版)
- Actions (VSports app下载)
- V体育ios版 - Actions
- Actions (VSports在线直播)
- "V体育安卓版" Actions
- "VSports在线直播" Actions
- Actions (V体育平台登录)
- Actions (V体育平台登录)
- Actions (V体育2025版)
Substances
- V体育安卓版 - Actions
- "V体育官网入口" Actions
- "VSports注册入口" Actions
- Actions (V体育安卓版)
- Actions (V体育平台登录)
- Actions (V体育官网)
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous (V体育安卓版)

