"VSports注册入口" Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish
- PMID: 21538441
- PMCID: "VSports注册入口" PMC3145024
- DOI: 10.1002/hep.24396
Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish
VSports手机版 - Abstract
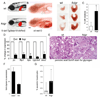
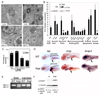
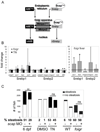
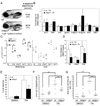
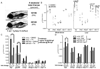
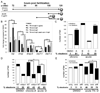
Many etiologies of fatty liver disease (FLD) are associated with the hyperactivation of one of the three pathways composing the unfolded protein response (UPR), which is a harbinger of endoplasmic reticulum (ER) stress. The UPR is mediated by pathways initiated by PRKR-like endoplasmic reticulum kinase, inositol-requiring 1A/X box binding protein 1, and activating transcription factor 6 (ATF6), and each of these pathways has been implicated to have a protective or pathological role in FLD. We used zebrafish with FLD and hepatic ER stress to explore the relationship between Atf6 and steatosis. A mutation of the foie gras (foigr) gene caused FLD and hepatic ER stress. The prolonged treatment of wild-type larvae with tunicamycin (TN), which caused chronic ER stress, phenocopied foigr. In contrast, acute exposure to a high dose of TN robustly activated the UPR but was less effective at inducing steatosis. The sterol regulatory element binding protein transcription factors were not required for steatosis in any of these models. Instead, depleting larvae of active Atf6 either through a membrane-bound transcription factor peptidase site 1 mutation or an atf6 morpholino injection protected them against steatosis caused by chronic ER stress, but exacerbated steatosis caused by acute TN treatment. VSports手机版.
Conclusion: ER stress causes FLD. A loss of Atf6 prevents steatosis caused by chronic ER stress but can also potentiate steatosis caused by acute ER stress V体育安卓版. This demonstrates that Atf6 can play both protective and pathological roles in FLD. .
Copyright © 2011 American Association for the Study of Liver Diseases. V体育ios版.
Figures


References
-
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. - PubMed
-
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. - "VSports app下载" PubMed
-
- Asselah T, Bieche I, Mansouri A, Laurendeau I, Cazals-Hatem D, Feldmann G, Bedossa P, et al. In vivo hepatic endoplasmic reticulum stress in patients with chronic hepatitis C. J Pathol. 2010;221:264–274. - PubMed
Publication types
- "VSports在线直播" Actions
MeSH terms
- V体育安卓版 - Actions
- Actions (VSports)
- Actions (V体育官网入口)
- Actions (VSports在线直播)
- Actions (VSports注册入口)
- "V体育官网入口" Actions
Substances
- V体育平台登录 - Actions
- VSports手机版 - Actions
- "VSports app下载" Actions
VSports最新版本 - Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
