VSports注册入口 - OTUbase: an R infrastructure package for operational taxonomic unit data
- PMID: 21498398
- PMCID: PMC3106189
- DOI: 10.1093/bioinformatics/btr196 (VSports app下载)
OTUbase: an R infrastructure package for operational taxonomic unit data (V体育平台登录)
Abstract
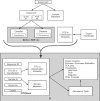
Summary: OTUbase is an R package designed to facilitate the analysis of operational taxonomic unit (OTU) data and sequence classification (taxonomic) data. Currently there are programs that will cluster sequence data into OTUs and/or classify sequence data into known taxonomies. However, there is a need for software that can take the summarized output of these programs and organize it into easily accessed and manipulated formats. OTUbase provides this structure and organization within R, to allow researchers to easily manipulate the data with the rich library of R packages currently available for additional analysis. VSports手机版.
Availability: OTUbase is an R package available through Bioconductor. It can be found at http://www. bioconductor. org/packages/release/bioc/html/OTUbase. html. V体育安卓版.
Figures
References
-
- Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. - VSports最新版本 - PMC - PubMed
-
- Giongo A, et al. PANGEA: pipeline for analysis of next generation amplicons. The ISME Journal. 2010;4:852–861. - "VSports app下载" PMC - PubMed
-
- Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:reviews0003.1–reviews0003.8. - PMC (V体育2025版) - PubMed
-
- Kunin V, et al. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 2010;12:118–123. - PubMed
Publication types
- "V体育平台登录" Actions
MeSH terms
- "V体育2025版" Actions
- V体育平台登录 - Actions
- "V体育官网入口" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

