Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells
- PMID: 21295524
- PMCID: "V体育平台登录" PMC3040415
- DOI: VSports app下载 - 10.1016/j.ccr.2010.12.022
Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells
Abstract (VSports手机版)
Despite evidence supporting an oncogenic role in breast cancer, the Notch pathway's contribution to metastasis remains unknown. Here, we report that the Notch ligand Jagged1 is a clinically and functionally important mediator of bone metastasis by activating the Notch pathway in bone cells VSports手机版. Jagged1 promotes tumor growth by stimulating IL-6 release from osteoblasts and directly activates osteoclast differentiation. Furthermore, Jagged1 is a potent downstream mediator of the bone metastasis cytokine TGFβ that is released during bone destruction. Importantly, γ-secretase inhibitor treatment reduces Jagged1-mediated bone metastasis by disrupting the Notch pathway in stromal bone cells. These findings elucidate a stroma-dependent mechanism for Notch signaling in breast cancer and provide rationale for using γ-secretase inhibitors for the treatment of bone metastasis. .
Copyright © 2011 Elsevier Inc. All rights reserved. V体育安卓版.
Figures



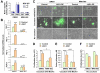
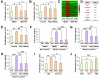
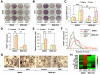


Comment in
-
One NOTCH Further: Jagged 1 in Bone Metastasis.Cancer Cell. 2011 Feb 15;19(2):159-61. doi: 10.1016/j.ccr.2011.01.043. Cancer Cell. 2011. PMID: 21316596
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science (New York, NY. 1999;284:770–776. - PubMed
-
- Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer research. 1992;52:1399–1405. - PubMed
-
- Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem. 2008;283:6509–6518. - V体育安卓版 - PubMed
-
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. - PubMed
Publication types
- VSports注册入口 - Actions
- "VSports手机版" Actions
MeSH terms
- Actions (VSports)
- "VSports" Actions
- Actions (V体育官网)
- V体育2025版 - Actions
- Actions (VSports app下载)
- Actions (V体育官网入口)
- V体育ios版 - Actions
Substances
- "VSports app下载" Actions
- "V体育2025版" Actions
- VSports - Actions
- "V体育ios版" Actions
- "V体育官网入口" Actions
- VSports手机版 - Actions
Associated data
- Actions
Grants and funding
LinkOut - more resources
"VSports app下载" Full Text Sources
"V体育2025版" Other Literature Sources
Medical
VSports注册入口 - Molecular Biology Databases

