More forks on the road to replication stress recovery
- PMID: 21278446
- PMCID: PMC3030971
- DOI: 10.1093/jmcb/mjq049 (VSports注册入口)
VSports手机版 - More forks on the road to replication stress recovery
Abstract
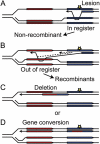
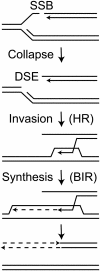
High-fidelity replication of DNA, and its accurate segregation to daughter cells, is critical for maintaining genome stability and suppressing cancer. DNA replication forks are stalled by many DNA lesions, activating checkpoint proteins that stabilize stalled forks. Stalled forks may eventually collapse, producing a broken DNA end. Fork restart is typically mediated by proteins initially identified by their roles in homologous recombination repair of DNA double-strand breaks (DSBs). In recent years, several proteins involved in DSB repair by non-homologous end joining (NHEJ) have been implicated in the replication stress response, including DNA-PKcs, Ku, DNA Ligase IV-XRCC4, Artemis, XLF and Metnase. It is currently unclear whether NHEJ proteins are involved in the replication stress response through indirect (signaling) roles, and/or direct roles involving DNA end joining. Additional complexity in the replication stress response centers around RPA, which undergoes significant post-translational modification after stress, and RAD52, a conserved HR protein whose role in DSB repair may have shifted to another protein in higher eukaryotes, such as BRCA2, but retained its role in fork restart. Most cancer therapeutic strategies create DNA replication stress. Thus, it is imperative to gain a better understanding of replication stress response proteins and pathways to improve cancer therapy. VSports手机版.
Figures

References
-
- Adachi N., So S., Koyama H. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 2004;279:37343–37348. doi:10.1074/jbc.M313910200. - DOI - PubMed
-
- Ahnesorg P., Smith P., Jackson S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124:301–313. doi:10.1016/j.cell.2005.12.031. - DOI - PubMed
-
- Allen C., Kurimasa A., Brennemann M.A., et al. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. USA. 2002;99:3758–3763. doi:10.1073/pnas.052545899. - DOI - PMC - PubMed
-
- Anantha R.W., Vassin V.M., Borowiec J.A. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. VSports - doi:10.1074/jbc.M704645200. - DOI - PubMed
-
- Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. V体育平台登录 - doi:10.1006/jmbi.2001.4564. - DOI - PubMed
Publication types
- "VSports最新版本" Actions
- VSports注册入口 - Actions
MeSH terms
- "VSports最新版本" Actions
- Actions (V体育2025版)
- "V体育ios版" Actions
- "V体育2025版" Actions
Substances
- "V体育2025版" Actions
- "VSports app下载" Actions
Grants and funding
LinkOut - more resources (V体育官网)
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

