Nonsense mediated decay resistant mutations are a source of expressed mutant proteins in colon cancer cell lines with microsatellite instability (VSports注册入口)
- PMID: 21209843
- PMCID: PMC3013145
- DOI: 10.1371/journal.pone.0016012
Nonsense mediated decay resistant mutations are a source of expressed mutant proteins in colon cancer cell lines with microsatellite instability
Erratum in
- PLoS One. 2011;6(1). doi: 10.1371/annotation/53805ecf-7d10-4d99-9cec-f27f5e0d4166. Walker, Franscesa [corrected to Walker, Francesca]
Abstract
Background: Frameshift mutations in microsatellite instability high (MSI-High) colorectal cancers are a potential source of targetable neo-antigens. Many nonsense transcripts are subject to rapid degradation due to nonsense-mediated decay (NMD), but nonsense transcripts with a cMS in the last exon or near the last exon-exon junction have intrinsic resistance to nonsense-mediated decay (NMD) VSports手机版. NMD-resistant transcripts are therefore a likely source of expressed mutant proteins in MSI-High tumours. .
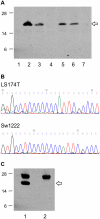
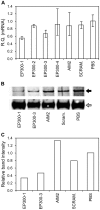
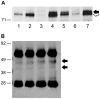
Methods: Using antibodies to the conserved N-termini of predicted mutant proteins, we analysed MSI-High colorectal cancer cell lines for examples of naturally expressed mutant proteins arising from frameshift mutations in coding microsatellites (cMS) by immunoprecipitation and Western Blot experiments. Detected mutant protein bands from NMD-resistant transcripts were further validated by gene-specific short-interfering RNA (siRNA) knockdown. A genome-wide search was performed to identify cMS-containing genes likely to generate NMD-resistant transcripts that could encode for antigenic expressed mutant proteins in MSI-High colon cancers. These genes were screened for cMS mutations in the MSI-High colon cancer cell lines V体育安卓版. .
Results: Mutant protein bands of expected molecular weight were detected in mutated MSI-High cell lines for NMD-resistant transcripts (CREBBP, EP300, TTK), but not NMD-sensitive transcripts (BAX, CASP5, MSH3). Expression of the mutant CREBBP and EP300 proteins was confirmed by siRNA knockdown. Five cMS-bearing genes identified from the genome-wide search and without existing mutation data (SFRS12IP1, MED8, ASXL1, FBXL3 and RGS12) were found to be mutated in at least 5 of 11 (45%) of the MSI-High cell lines tested. V体育ios版.
Conclusion: NMD-resistant transcripts can give rise to expressed mutant proteins in MSI-High colon cancer cells. If commonly expressed in primary MSI-High colon cancers, MSI-derived mutant proteins could be useful as cancer specific immunological targets in a vaccine targeting MSI-High colonic tumours. VSports最新版本.
VSports手机版 - Conflict of interest statement
Figures


References
-
- Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, et al. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative Real Common Target genes. Oncogene. 2003;22:2226–2235. - PubMed
-
- Duval A, Hamelin R. Genetic instability in human mismatch repair deficient cancers. Ann Genet. 2002;45:71–75. - PubMed
-
- Niv Y. Microsatellite instability and MLH1 promoter hypermethylation in colorectal cancer. World J Gastroenterol. 2007;13:1767–1769. - PMC (V体育平台登录) - PubMed
-
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. - "V体育官网" PubMed
-
- Jenkins MA, Hayashi S, O'Shea AM, Burgart LJ, Smyrk TC, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. - VSports - PMC - PubMed
Publication types
"V体育ios版" MeSH terms
- VSports最新版本 - Actions
- Actions (VSports在线直播)
- VSports app下载 - Actions
- VSports app下载 - Actions
- Actions (V体育官网)
- VSports app下载 - Actions
- V体育官网入口 - Actions
- "VSports在线直播" Actions
Substances
- Actions (VSports在线直播)
LinkOut - more resources
Full Text Sources (VSports最新版本)
Other Literature Sources
Molecular Biology Databases (VSports最新版本)
Research Materials
Miscellaneous

