V体育2025版 - Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection
- PMID: 21083371
- PMCID: PMC3107555
- DOI: 10.1086/657322 (VSports)
Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection (V体育2025版)
Abstract
Background: High-throughput genome-wide techniques have facilitated the identification of previously unknown host proteins involved in cellular human immunodeficiency virus (HIV) infection. Recently, 3 independent studies have used small interfering RNA technology to silence each gene in the human genome to determine the importance of each in HIV infection. Genes conferring a significant effect were termed HIV-dependency factors (HDFs). VSports手机版.
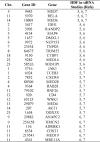
Methods: We assembled high-density panels of 6380 single-nucleotide polymorphisms (SNPs) in 278 HDF genes and tested for genotype associations with HIV infection and AIDS progression in 1633 individuals from clinical AIDS cohorts. V体育安卓版.
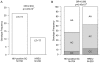
Results: After statistical correction for multiple tests, significant associations with HIV acquisition were found for SNPs in 2 genes, NCOR2 and IDH1. Weaker associations with AIDS progression were revealed for SNPs within the TM9SF2 and EGFR genes V体育ios版. .
Conclusions: This study independently verifies the influence of NCOR2 and IDH1 on HIV transmission, and its findings suggest that variation in these genes affects susceptibility to HIV infection in exposed individuals VSports最新版本. .
Figures









References (VSports手机版)
-
- Gallo RC, Montagnier L. The discovery of HIV as the cause of AIDS. N Engl J Med. 2003;349:2283–2285. - PubMed
-
- Vandekerckhove L, Verhofstede C, Vogelaers D. Maraviroc: integration of a new antiretroviral drug class into clinical practice. J Antimicrob Chemother. 2008;61:1187–1190. - "VSports在线直播" PubMed
-
- Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV- 1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. - PubMed (V体育官网)
-
- Menendez-Arias L. Molecular basis of human immunodeficiency virus drug resistance: an update. Antiviral Res. 2010;85(1):210–231. - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. - PubMed
Publication types
MeSH terms
- V体育官网 - Actions
- "VSports最新版本" Actions
- VSports - Actions
- Actions (V体育官网入口)
Substances
- V体育安卓版 - Actions
- VSports注册入口 - Actions
Grants and funding
- UO1‐AI‐37613/AI/NIAID NIH HHS/United States
- VSports最新版本 - N02-CP‐55504/CP/NCI NIH HHS/United States
- "V体育ios版" R56 DA004334/DA/NIDA NIH HHS/United States
- N01‐CO‐12400/CO/NCI NIH HHS/United States
- R01 DA004334/DA/NIDA NIH HHS/United States (VSports最新版本)
- UO1‐AI‐35039/AI/NIAID NIH HHS/United States
- U01 AI037613/AI/NIAID NIH HHS/United States (VSports注册入口)
- U01 AI035041/AI/NIAID NIH HHS/United States
- UO1‐AI‐35040/AI/NIAID NIH HHS/United States
- N01 CO012400/CA/NCI NIH HHS/United States (V体育官网入口)
- R01 HD041224/HD/NICHD NIH HHS/United States
- UO1‐AI‐35041/AI/NIAID NIH HHS/United States
- "VSports" N02 CP055504/CP/NCI NIH HHS/United States
- U01 AI035040/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- 1 R01 HD41224/HD/NICHD NIH HHS/United States
- UO1‐AI‐35043/AI/NIAID NIH HHS/United States (VSports app下载)
- "V体育ios版" U01 AI035042/AI/NIAID NIH HHS/United States
- M01 RR00425/RR/NCRR NIH HHS/United States
- U01 AI037984/AI/NIAID NIH HHS/United States
- UO1‐AI‐37984/AI/NIAID NIH HHS/United States
- "VSports手机版" R01‐DA‐04334/DA/NIDA NIH HHS/United States
- "VSports app下载" M01 RR000722/RR/NCRR NIH HHS/United States
- 5‐M01‐RR‐00722/RR/NCRR NIH HHS/United States (VSports)
- M01 RR000425/RR/NCRR NIH HHS/United States
- U01 AI035043/AI/NIAID NIH HHS/United States
- N01 CO012400/CO/NCI NIH HHS/United States (V体育安卓版)
- R01 DA012568/DA/NIDA NIH HHS/United States
- UO1‐AI‐35042/AI/NIAID NIH HHS/United States
- U01 AI035039/AI/NIAID NIH HHS/United States
- R01‐DA‐12568/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
VSports最新版本 - Miscellaneous

