Mitochondrial DNA replication and disease: insights from DNA polymerase γ mutations
- PMID: 20927567
- PMCID: "VSports注册入口" PMC3046768
- DOI: 10.1007/s00018-010-0530-4 (V体育官网)
Mitochondrial DNA replication and disease: insights from DNA polymerase γ mutations
Abstract
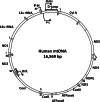
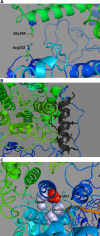
DNA polymerase γ (pol γ), encoded by POLG, is responsible for replicating human mitochondrial DNA. About 150 mutations in the human POLG have been identified in patients with mitochondrial diseases such as Alpers syndrome, progressive external ophthalmoplegia, and ataxia-neuropathy syndromes VSports手机版. Because many of the mutations are described in single citations with no genotypic family history, it is important to ascertain which mutations cause or contribute to mitochondrial disease. The vast majority of data about POLG mutations has been generated from biochemical characterizations of recombinant pol γ. However, recently, the study of mitochondrial dysfunction in Saccharomyces cerevisiae and mouse models provides important in vivo evidence for the role of POLG mutations in disease. Also, the published 3D-structure of the human pol γ assists in explaining some of the biochemical and genetic properties of the mutants. This review summarizes the current evidence that identifies and explains disease-causing POLG mutations. .
Figures


VSports手机版 - References
-
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. - PubMed
-
- Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. - PubMed
-
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. - VSports - PubMed
-
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106:383–405. - VSports手机版 - PubMed
-
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. - PubMed
V体育官网 - Publication types
- Actions (VSports)
- "V体育ios版" Actions
MeSH terms
- "V体育官网入口" Actions
- VSports - Actions
- "VSports app下载" Actions
- "VSports在线直播" Actions
- VSports最新版本 - Actions
- V体育ios版 - Actions
- "V体育平台登录" Actions
- V体育安卓版 - Actions
- Actions (VSports在线直播)
"V体育安卓版" Substances
- Actions (V体育ios版)
Grants and funding
LinkOut - more resources
Full Text Sources
"VSports最新版本" Other Literature Sources
VSports在线直播 - Medical
Molecular Biology Databases

