Orphan CpG islands identify numerous conserved promoters in the mammalian genome
- PMID: 20885785
- PMCID: PMC2944787
- DOI: 10.1371/journal.pgen.1001134
"VSports最新版本" Orphan CpG islands identify numerous conserved promoters in the mammalian genome
V体育平台登录 - Abstract
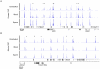
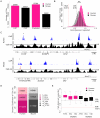
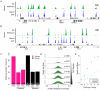
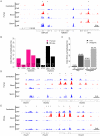
CpG islands (CGIs) are vertebrate genomic landmarks that encompass the promoters of most genes and often lack DNA methylation. Querying their apparent importance, the number of CGIs is reported to vary widely in different species and many do not co-localise with annotated promoters. We set out to quantify the number of CGIs in mouse and human genomes using CXXC Affinity Purification plus deep sequencing (CAP-seq). We also asked whether CGIs not associated with annotated transcripts share properties with those at known promoters. We found that, contrary to previous estimates, CGI abundance in humans and mice is very similar and many are at conserved locations relative to genes. In each species CpG density correlates positively with the degree of H3K4 trimethylation, supporting the hypothesis that these two properties are mechanistically interdependent. Approximately half of mammalian CGIs (>10,000) are "orphans" that are not associated with annotated promoters VSports手机版. Many orphan CGIs show evidence of transcriptional initiation and dynamic expression during development. Unlike CGIs at known promoters, orphan CGIs are frequently subject to DNA methylation during development, and this is accompanied by loss of their active promoter features. In colorectal tumors, however, orphan CGIs are not preferentially methylated, suggesting that cancer does not recapitulate a developmental program. Human and mouse genomes have similar numbers of CGIs, over half of which are remote from known promoters. Orphan CGIs nevertheless have the characteristics of functional promoters, though they are much more likely than promoter CGIs to become methylated during development and hence lose these properties. The data indicate that orphan CGIs correspond to previously undetected promoters whose transcriptional activity may play a functional role during development. .
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - "V体育2025版" PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - "V体育2025版" PubMed
-
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. - PubMed
-
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. - PubMed
Publication types
MeSH terms
- "VSports" Actions
- Actions (V体育官网入口)
- Actions (VSports)
- Actions (VSports)
- "V体育官网入口" Actions
- "V体育2025版" Actions
- "VSports在线直播" Actions
Substances
- V体育ios版 - Actions
- "V体育ios版" Actions
Grants and funding
LinkOut - more resources
Full Text Sources (V体育2025版)
Other Literature Sources
Molecular Biology Databases
Miscellaneous

