A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response
- PMID: 20673990
- PMCID: PMC2956184
- DOI: 10.1016/j.cell.2010.06.040
A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response
"VSports注册入口" Abstract
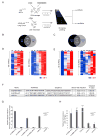
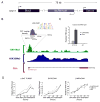
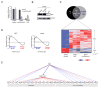
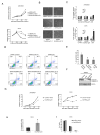
Recently, more than 1000 large intergenic noncoding RNAs (lincRNAs) have been reported. These RNAs are evolutionarily conserved in mammalian genomes and thus presumably function in diverse biological processes VSports手机版. Here, we report the identification of lincRNAs that are regulated by p53. One of these lincRNAs (lincRNA-p21) serves as a repressor in p53-dependent transcriptional responses. Inhibition of lincRNA-p21 affects the expression of hundreds of gene targets enriched for genes normally repressed by p53. The observed transcriptional repression by lincRNA-p21 is mediated through the physical association with hnRNP-K. This interaction is required for proper genomic localization of hnRNP-K at repressed genes and regulation of p53 mediates apoptosis. We propose a model whereby transcription factors activate lincRNAs that serve as key repressors by physically associating with repressive complexes and modulate their localization to sets of previously active genes. .
Copyright 2010 Elsevier Inc V体育安卓版. All rights reserved. .
Figures (V体育平台登录)
"VSports app下载" Comment in
-
"VSports" Noncoding RNAs: the missing "linc" in p53-mediated repression.Cell. 2010 Aug 6;142(3):358-60. doi: 10.1016/j.cell.2010.07.029. Cell. 2010. PMID: 20691894
References
-
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. - PubMed
-
- Carninci P. Non-coding RNA transcription: turning on neighbours. Nat Cell Biol. 2008;10:1023–1024. - PubMed
-
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. - PubMed
Publication types
MeSH terms
- "V体育官网" Actions
- "VSports在线直播" Actions
- VSports在线直播 - Actions
- "VSports在线直播" Actions
- "VSports在线直播" Actions
Substances
- Actions (V体育2025版)
- Actions (VSports手机版)
- "VSports手机版" Actions
V体育官网入口 - Associated data
- Actions (V体育平台登录)
Grants and funding
LinkOut - more resources
Full Text Sources
"V体育官网入口" Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

