Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis
- PMID: 20660630
- PMCID: "VSports注册入口" PMC2930282
- DOI: 10.1083/jcb.201001129 (VSports)
"V体育2025版" Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis
Abstract
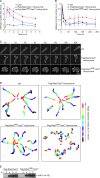
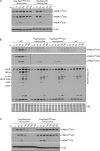
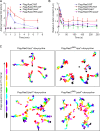
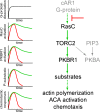
In chemotactic cells, G protein-coupled receptors activate Ras proteins, but it is unclear how Ras-associated pathways link extracellular signaling to cell migration VSports手机版. We show that, in Dictyostelium discoideum, activated forms of RasC prolong the time course of TORC2 (target of rapamycin [Tor] complex 2)-mediated activation of a myristoylated protein kinase B (PKB; PKBR1) and the phosphorylation of PKB substrates, independently of phosphatidylinositol-(3,4,5)-trisphosphate. Paralleling these changes, the kinetics of chemoattractant-induced adenylyl cyclase activation and actin polymerization are extended, pseudopodial activity is increased and mislocalized, and chemotaxis is impaired. The effects of activated RasC are suppressed by deletion of the TORC2 subunit PiaA. In vitro RasC(Q62L)-dependent PKB phosphorylation can be rapidly initiated by the addition of a PiaA-associated immunocomplex to membranes of TORC2-deficient cells and blocked by TOR-specific inhibitor PP242. Furthermore, TORC2 binds specifically to the activated form of RasC. These results demonstrate that RasC is an upstream regulator of TORC2 and that the TORC2-PKB signaling mediates effects of activated Ras proteins on the cytoskeleton and cell migration. .
Figures





VSports app下载 - References
-
- Cantley L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657 10.1126/science.296.5573.1655 - DOI (VSports手机版) - PubMed
-
- Feig L.A., Cooper G.M. 1988. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell. Biol. 8:2472–2478 - PMC (VSports最新版本) - PubMed
"VSports app下载" Publication types
- Actions (V体育ios版)
- Actions (V体育2025版)
MeSH terms
- Actions (VSports)
- Actions (V体育平台登录)
- Actions (VSports注册入口)
- Actions (VSports在线直播)
- "VSports注册入口" Actions
- V体育2025版 - Actions
- "VSports app下载" Actions
Substances
- "V体育平台登录" Actions
- "V体育平台登录" Actions
Grants and funding
LinkOut - more resources
"VSports最新版本" Full Text Sources
Other Literature Sources
Molecular Biology Databases
V体育2025版 - Research Materials
"VSports" Miscellaneous

