A boundary delimitation algorithm to approximate cell soma volumes of bipolar cells from topographical data obtained by scanning probe microscopy (V体育安卓版)
- PMID: 20550692
- PMCID: PMC2912302
- DOI: V体育安卓版 - 10.1186/1471-2105-11-323
A boundary delimitation algorithm to approximate cell soma volumes of bipolar cells from topographical data obtained by scanning probe microscopy
Abstract
Background: Cell volume determination plays a pivotal role in the investigation of the biophysical mechanisms underlying various cellular processes. Whereas light microscopy in principle enables one to obtain three dimensional data, the reconstruction of cell volume from z-stacks is a time consuming procedure. Thus, three dimensional topographic representations of cells are easier to obtain by scanning probe microscopical measurements VSports手机版. .
Results: We present a method of separating the cell soma volume of bipolar cells in adherent cell cultures from the contributions of the cell processes from data obtained by scanning ion conductance microscopy. Soma volume changes between successive scans obtained from the same cell can then be computed even if the cell is changing its position within the observed area. We demonstrate that the estimation of the cell volume on the basis of the width and the length of a cell may lead to erroneous determination of cell volume changes. V体育安卓版.
Conclusions: We provide a new algorithm to repeatedly determine single cell soma volume and thus to quantify cell volume changes during cell movements occuring over a time range of hours. V体育ios版.
Figures

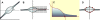

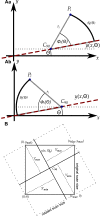

 of the rotated data and its surrounding four projections of the original data,
of the rotated data and its surrounding four projections of the original data,  to
to  The triangles Mk consisting of three of the projections
The triangles Mk consisting of three of the projections  are indicated by the dotted lines. D: The rotated data set with C90 located in the origin. C: The sum ζk of the three angles at
are indicated by the dotted lines. D: The rotated data set with C90 located in the origin. C: The sum ζk of the three angles at  to the three points of a triangle is 2π.
to the three points of a triangle is 2π.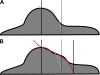
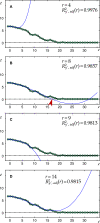
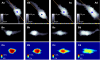
 were compared. Since
were compared. Since  Xy' (r = 8) (red arrow-head in B) was selected as the boundary of the cell soma for the investigated y'.
Xy' (r = 8) (red arrow-head in B) was selected as the boundary of the cell soma for the investigated y'.
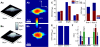
 Note that the chart omits some additional tests (see text) to ensure an error free operation as indicated by the dotted arrow in the lower right part. Note that NaN ≡ not a number.
Note that the chart omits some additional tests (see text) to ensure an error free operation as indicated by the dotted arrow in the lower right part. Note that NaN ≡ not a number.

"VSports app下载" References
-
- Lang F, Ritter M, Völkl H, Häussinger D. The biological significance of cell volume. Ren Physiol Biochem. 1993;11(12):48–65. - PubMed
-
- Wehner F, Olsen H, Tinel H, Kinne-Saffran E, Kinne RKH. Cell volume regulation: osmolytes, osmolyte transport, and signal transduction. Rev Physiol Biochem Pharmacol. 2003;11:1–80. full_text. - "V体育官网入口" PubMed
-
- Hall SK, Zhang J, Lieberman M. An early transient current is associated with hyposmotic swelling and volume regulation in embryonic chick cardiac myocytes. Exp Physiol. 1997;11:43–54. - "VSports" PubMed
V体育官网 - Publication types
- Actions (VSports)
MeSH terms
- Actions (V体育官网入口)
- VSports手机版 - Actions
- "V体育平台登录" Actions
- "VSports手机版" Actions
- "V体育官网" Actions
V体育官网 - LinkOut - more resources
Full Text Sources
Medical

