Frataxin and mitochondrial FeS cluster biogenesis
- PMID: 20522547
- PMCID: VSports app下载 - PMC2930671
- DOI: 10.1074/jbc.R110.118679
"VSports" Frataxin and mitochondrial FeS cluster biogenesis
Abstract
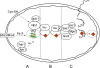
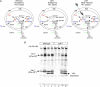
Friedreich ataxia is an inherited neurodegenerative disease caused by frataxin deficiency. Frataxin is a conserved mitochondrial protein that plays a role in FeS cluster assembly in mitochondria. FeS clusters are modular cofactors that perform essential functions throughout the cell. They are synthesized by a multistep and multisubunit mitochondrial machinery that includes the scaffold protein Isu for assembling a protein-bound FeS cluster intermediate VSports手机版. Frataxin interacts with Isu, iron, and the cysteine desulfurase Nfs1, which supplies sulfide, thus placing it at the center of mitochondrial FeS cluster biosynthesis. .
Figures (VSports注册入口)
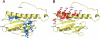
References
-
- Pandolfo M. (2009) J. Neurol. 256, Suppl. 1, 3–8 - PubMed
-
- Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P. I., Di Donato S., Mandel J. L., Cocozza S., Koenig M., Pandolfo M. (1996) Science 271, 1423–1427 - VSports手机版 - PubMed
-
- Wilson R. B., Roof D. M. (1997) Nat. Genet. 16, 352–357 - "VSports注册入口" PubMed
-
- Campuzano V., Montermini L., Lutz Y., Cova L., Hindelang C., Jiralerspong S., Trottier Y., Kish S. J., Faucheux B., Trouillas P., Authier F. J., Dürr A., Mandel J. L., Vescovi A., Pandolfo M., Koenig M. (1997) Hum. Mol. Genet. 6, 1771–1780 - PubMed
-
- Gordon D. M., Shi Q., Dancis A., Pain D. (1999) Hum. Mol. Genet. 8, 2255–2262 - PubMed
Publication types
MeSH terms
- V体育官网 - Actions
- Actions (V体育安卓版)
- VSports最新版本 - Actions
- "V体育ios版" Actions
- "V体育安卓版" Actions
- V体育ios版 - Actions
- Actions (V体育安卓版)
- "VSports在线直播" Actions
Substances
- Actions (VSports app下载)
Grants and funding
LinkOut - more resources
Full Text Sources (VSports注册入口)
Other Literature Sources
Medical (V体育官网)
Molecular Biology Databases
Miscellaneous

