Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression
- PMID: 20386740
- PMCID: PMC2851566
- DOI: 10.1371/journal.pgen.1000899 (VSports注册入口)
Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression (VSports)
Abstract
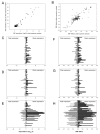
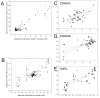
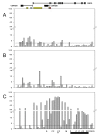
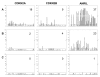
Single nucleotide polymorphisms (SNPs) on chromosome 9p21 are associated with coronary artery disease, diabetes, and multiple cancers. Risk SNPs are mainly non-coding, suggesting that they influence expression and may act in cis. We examined the association between 56 SNPs in this region and peripheral blood expression of the three nearest genes CDKN2A, CDKN2B, and ANRIL using total and allelic expression in two populations of healthy volunteers: 177 British Caucasians and 310 mixed-ancestry South Africans. Total expression of the three genes was correlated (P<0. 05), suggesting that they are co-regulated. SNP associations mapped by allelic and total expression were similar (r = 0. 97, P = 4. 8x10(-99)), but the power to detect effects was greater for allelic expression. The proportion of expression variance attributable to cis-acting effects was 8% for CDKN2A, 5% for CDKN2B, and 20% for ANRIL. SNP associations were similar in the two populations (r = 0. 94, P = 10(-72)). Multiple SNPs were independently associated with expression of each gene (P<0 VSports手机版. 05 after correction for multiple testing), suggesting that several sites may modulate disease susceptibility. Individual SNPs correlated with changes in expression up to 1. 4-fold for CDKN2A, 1. 3-fold for CDKN2B, and 2-fold for ANRIL. Risk SNPs for coronary disease, stroke, diabetes, melanoma, and glioma were all associated with allelic expression of ANRIL (all P<0. 05 after correction for multiple testing), while association with the other two genes was only detectable for some risk SNPs. SNPs had an inverse effect on ANRIL and CDKN2B expression, supporting a role of antisense transcription in CDKN2B regulation. Our study suggests that modulation of ANRIL expression mediates susceptibility to several important human diseases. .
V体育安卓版 - Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


V体育ios版 - References
-
- Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, et al. A Common Variant on Chromosome 9p21 Affects the Risk of Myocardial Infarction. Science. 2007;316:1491–1493. - PubMed
Publication types
- "V体育平台登录" Actions
MeSH terms
- VSports最新版本 - Actions
- VSports注册入口 - Actions
- "V体育2025版" Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
- "VSports app下载" Actions
- V体育安卓版 - Actions
- "VSports手机版" Actions
- Actions (VSports app下载)
Substances
Grants and funding
LinkOut - more resources
Full Text Sources (VSports注册入口)
Other Literature Sources
Miscellaneous

