vttRA and vttRB Encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain
- PMID: 20385759
- PMCID: PMC2876560
- DOI: 10.1128/IAI.01073-09
vttRA and vttRB Encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain (VSports)
Abstract
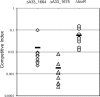
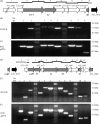
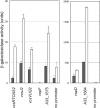
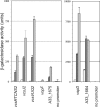
Strain AM-19226 is a pathogenic non-O1/non-O139 serogroup Vibrio cholerae strain that does not encode the toxin-coregulated pilus or cholera toxin but instead causes disease using a type three secretion system (T3SS). Two genes within the T3SS pathogenicity island, herein named vttR(A) (locus tag A33_1664) and vttR(B) (locus tag A33_1675), are predicted to encode proteins that show similarity to the transcriptional regulator ToxR, which is found in all strains of V. cholerae. Strains with a deletion of vttR(A) or vttR(B) showed attenuated colonization in vivo, indicating that the T3SS-encoded regulatory proteins play a role in virulence. lacZ transcriptional reporter fusions to intergenic regions upstream of genes encoding the T3SS structural components identified growth in the presence of bile as a condition that modulates gene expression. Under this condition, VttR(A) and VttR(B) were necessary for maximal gene expression. In contrast, growth in bile did not substantially alter the expression of a reporter fusion to the vopF gene, which encodes an effector protein. Increased vttR(B) reporter fusion activity was observed in a DeltavttR(B) strain background, suggesting that VttR(B) may regulate its own expression. The collective results are consistent with the hypothesis that T3SS-encoded regulatory proteins are essential for pathogenesis and control the expression of selected T3SS genes VSports手机版. .
Figures

References
-
- Alam, A., R. C. Larocque, J. B. Harris, C. Vanderspurt, E. T. Ryan, F. Qadri, and S. B. Calderwood. 2005. Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse. Infect. Immun. 73:6674-6679. - "VSports注册入口" PMC - PubMed
-
- Anderson, A. M., J. B. Varkey, C. A. Petti, R. A. Liddle, R. Frothingham, and C. W. Woods. 2004. Non-O1 Vibrio cholerae septicemia: case report, discussion of literature, and relevance to bioterrorism. Diagn. Microbiol. Infect. Dis. 49:295-297. - PubMed (VSports最新版本)
Publication types
- "VSports在线直播" Actions
MeSH terms (VSports)
- Actions (VSports注册入口)
- "V体育ios版" Actions
- "V体育官网入口" Actions
- "VSports注册入口" Actions
- V体育官网 - Actions
Substances
- "V体育2025版" Actions
Grants and funding
LinkOut - more resources (V体育2025版)
Full Text Sources
Medical (VSports app下载)

