Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains
- PMID: 20308326
- PMCID: PMC2863704
- DOI: 10.1128/MCB.01046-09
Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains
Abstract
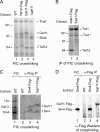
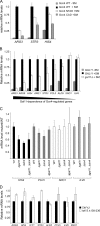
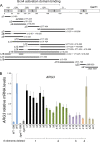
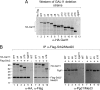
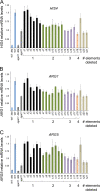
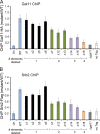
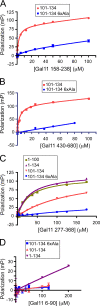
Targets of the tandem Gcn4 acidic activation domains in transcription preinitiation complexes were identified by site-specific cross-linking. The individual Gcn4 activation domains cross-link to three common targets, Gal11/Med15, Taf12, and Tra1, which are subunits of four conserved coactivator complexes, Mediator, SAGA, TFIID, and NuA4. The Gcn4 N-terminal activation domain also cross-links to the Mediator subunit Sin4/Med16. The contribution of the two Gcn4 activation domains to transcription was gene specific and varied from synergistic to less than additive VSports手机版. Gcn4-dependent genes had a requirement for Gal11 ranging from 10-fold dependence to complete Gal11 independence, while the Gcn4-Taf12 interaction did not significantly contribute to the expression of any gene studied. Complementary methods identified three conserved Gal11 activator-binding domains that bind each Gcn4 activation domain with micromolar affinity. These Gal11 activator-binding domains contribute additively to transcription activation and Mediator recruitment at Gcn4- and Gal11-dependent genes. Although we found that the conserved Gal11 KIX domain contributes to Gal11 function, we found no evidence of specific Gcn4-KIX interaction and conclude that the Gal11 KIX domain does not function by specific interaction with Gcn4. Our combined results show gene-specific coactivator requirements, a surprising redundancy in activator-target interactions, and an activator-coactivator interaction mediated by multiple low-affinity protein-protein interactions. .
Figures



References
-
- Altschuler, E. L., N. V. Hud, J. A. Mazrimas, and B. Rupp. 2000. Structure of polyglutamine. FEBS Lett. 472:166-168. - PubMed
-
- Barberis, A., J. Pearlberg, N. Simkovich, S. Farrell, P. Reinagel, C. Bamdad, G. Sigal, and M. Ptashne. 1995. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell 81:359-368. - PubMed
-
- Bhaumik, S. R., T. Raha, D. P. Aiello, and M. R. Green. 2004. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 18:333-343. - "VSports在线直播" PMC - PubMed
-
- Bochkareva, E., L. Kaustov, A. Ayed, G. S. Yi, Y. Lu, A. Pineda-Lucena, J. C. Liao, A. L. Okorokov, J. Milner, C. H. Arrowsmith, and A. Bochkarev. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. U. S. A. 102:15412-15417. - PMC - PubMed
-
- Bourbon, H. M. 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36:3993-4008. - "V体育安卓版" PMC - PubMed
"V体育官网入口" Publication types
V体育ios版 - MeSH terms
- "VSports app下载" Actions
- V体育官网 - Actions
- "VSports在线直播" Actions
- "V体育2025版" Actions
- "V体育安卓版" Actions
- Actions (VSports注册入口)
- VSports手机版 - Actions
- "VSports" Actions
- "VSports在线直播" Actions
- "VSports手机版" Actions
- Actions (VSports在线直播)
- VSports - Actions
- "VSports在线直播" Actions
- Actions (VSports注册入口)
Substances (V体育官网入口)
- "VSports最新版本" Actions
- Actions (VSports注册入口)
- Actions (VSports app下载)
- Actions (VSports手机版)
- V体育安卓版 - Actions
- "V体育2025版" Actions
Grants and funding (V体育官网)
LinkOut - more resources (VSports手机版)
Full Text Sources
"VSports" Other Literature Sources
Molecular Biology Databases (V体育平台登录)
Miscellaneous
