"VSports在线直播" Regulation of cellular metabolism by protein lysine acetylation
- PMID: 20167786
- PMCID: PMC3232675
- DOI: 10.1126/science.1179689
Regulation of cellular metabolism by protein lysine acetylation
Abstract
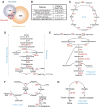
Protein lysine acetylation has emerged as a key posttranslational modification in cellular regulation, in particular through the modification of histones and nuclear transcription regulators. We show that lysine acetylation is a prevalent modification in enzymes that catalyze intermediate metabolism. Virtually every enzyme in glycolysis, gluconeogenesis, the tricarboxylic acid (TCA) cycle, the urea cycle, fatty acid metabolism, and glycogen metabolism was found to be acetylated in human liver tissue. The concentration of metabolic fuels, such as glucose, amino acids, and fatty acids, influenced the acetylation status of metabolic enzymes. Acetylation activated enoyl-coenzyme A hydratase/3-hydroxyacyl-coenzyme A dehydrogenase in fatty acid oxidation and malate dehydrogenase in the TCA cycle, inhibited argininosuccinate lyase in the urea cycle, and destabilized phosphoenolpyruvate carboxykinase in gluconeogenesis. Our study reveals that acetylation plays a major role in metabolic regulation VSports手机版. .
VSports最新版本 - Figures



Comment in
-
Cell biology. Rise of the rival.Science. 2010 Feb 19;327(5968):964-5. doi: 10.1126/science.1187159. Science. 2010. PMID: 20167774 No abstract available.
References
Publication types
MeSH terms
- "V体育ios版" Actions
- Actions (V体育官网)
- Actions (VSports)
- V体育ios版 - Actions
- V体育官网 - Actions
- VSports注册入口 - Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
- "V体育ios版" Actions
- Actions (V体育平台登录)
- "V体育ios版" Actions
- Actions (V体育官网)
- V体育官网 - Actions
- "V体育官网" Actions
- "V体育平台登录" Actions
- VSports在线直播 - Actions
- Actions (VSports最新版本)
Substances (V体育ios版)
- VSports手机版 - Actions
- Actions (VSports)
- Actions (V体育ios版)
- "VSports最新版本" Actions
- Actions (VSports app下载)
- Actions (V体育官网)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

