Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity
- PMID: 20144948
- PMCID: PMC2879524 (V体育ios版)
- DOI: 10.1093/nar/gkq048
Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity (VSports)
"VSports" Abstract
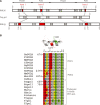
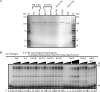
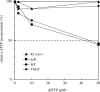
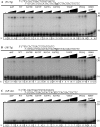
POLN is a nuclear A-family DNA polymerase encoded in vertebrate genomes. POLN has unusual fidelity and DNA lesion bypass properties, including strong strand displacement activity, low fidelity favoring incorporation of T for template G and accurate translesion synthesis past a 5S-thymine glycol (5S-Tg) VSports手机版. We searched for conserved features of the polymerase domain that distinguish it from prokaryotic pol I-type DNA polymerases. A Lys residue (679 in human POLN) of particular interest was identified in the conserved 'O-helix' of motif 4 in the fingers sub-domain. The corresponding residue is one of the most important for controlling fidelity of prokaryotic pol I and is a nonpolar Ala or Thr in those enzymes. Kinetic measurements show that K679A or K679T POLN mutant DNA polymerases have full activity on nondamaged templates, but poorly incorporate T opposite template G and do not bypass 5S-Tg efficiently. We also found that a conserved Tyr residue in the same motif not only affects sensitivity to dideoxynucleotides, but also greatly influences enzyme activity, fidelity and bypass. Protein sequence alignment reveals that POLN has three specific insertions in the DNA polymerase domain. The results demonstrate that residues have been strictly retained during evolution that confer unique bypass and fidelity properties on POLN. .
Figures (V体育ios版)

References
-
- Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low-fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 2006;281:23445–23455. - PubMed (VSports注册入口)
-
- Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase ν. DNA Repair. 2007;6:213–223. - PMC (VSports最新版本) - PubMed
-
- Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. - V体育官网入口 - PMC - PubMed
Publication types
- Actions (VSports在线直播)
- Actions (V体育2025版)
VSports app下载 - MeSH terms
- Actions (VSports最新版本)
- VSports最新版本 - Actions
- "VSports手机版" Actions
- Actions (VSports手机版)
Substances
- "V体育ios版" Actions
- "V体育平台登录" Actions
- "V体育ios版" Actions
- "VSports app下载" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials (VSports注册入口)
Miscellaneous

