V体育官网入口 - Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex
- PMID: 20081848
- PMCID: PMC2825759
- DOI: 10.1038/ni.1839
Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex
Abstract
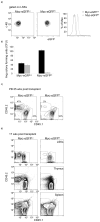
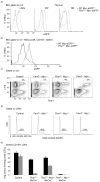
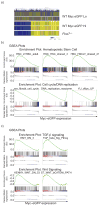
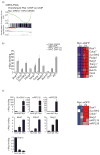
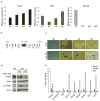
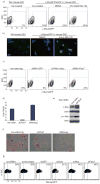
Hematopoietic stem cell (HSC) differentiation is regulated by cell-intrinsic and cell-extrinsic cues. In addition to transcriptional regulation, post-translational regulation may also control HSC differentiation. To test this hypothesis, we visualized the ubiquitin-regulated protein stability of a single transcription factor, c-Myc. The stability of c-Myc protein was indicative of HSC quiescence, and c-Myc protein abundance was controlled by the ubiquitin ligase Fbw7. Fine changes in the stability of c-Myc protein regulated the HSC gene-expression signature. Using whole-genome genomic approaches, we identified specific regulators of HSC function directly controlled by c-Myc binding; however, adult HSCs and embryonic stem cells sensed and interpreted c-Myc-regulated gene expression in distinct ways. Our studies show that a ubiquitin ligase-substrate pair can orchestrate the molecular program of HSC differentiation VSports手机版. .
Figures


Comment in
-
Controlling stem cell fate one substrate at a time.Nat Immunol. 2010 Mar;11(3):193-4. doi: 10.1038/ni0310-193. Nat Immunol. 2010. PMID: 20157300 No abstract available.
References
-
- Adams GB, Scadden DT. The hematopoietic stem cell in its place. Nat Immunol. 2006;7:333–337. - PubMed
-
- Kiel MJ, Morrison SJ. Uncertainty in the niches that maintain haematopoietic stem cells. Nat Rev Immunol. 2008;8:290–301. - PubMed
-
- Kondo M, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. - PubMed
-
- Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. - PubMed
Publication types
- "VSports最新版本" Actions
- "VSports手机版" Actions
MeSH terms
- Actions (V体育2025版)
- "V体育2025版" Actions
- Actions (VSports)
- "V体育2025版" Actions
- V体育官网入口 - Actions
- "V体育平台登录" Actions
- Actions (V体育2025版)
"VSports最新版本" Substances
- "V体育安卓版" Actions
- V体育平台登录 - Actions
Grants and funding
- T32 CA009161/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 CA133379/CA/NCI NIH HHS/United States
- R56AI070310/AI/NIAID NIH HHS/United States
- R01CA133379/CA/NCI NIH HHS/United States
- R21CA141399/CA/NCI NIH HHS/United States
- V体育官网入口 - R01 CA105129/CA/NCI NIH HHS/United States
- R01AI072039/AI/NIAID NIH HHS/United States
- R01 AI072039/AI/NIAID NIH HHS/United States
- R21 CA141399/CA/NCI NIH HHS/United States
- V体育ios版 - R01 AI041428/AI/NIAID NIH HHS/United States
- R01AI41428/AI/NIAID NIH HHS/United States
- R01CA105129/CA/NCI NIH HHS/United States (VSports注册入口)
- HHMI/Howard Hughes Medical Institute/United States
- R01CA120196/CA/NCI NIH HHS/United States
- R01 CA120196/CA/NCI NIH HHS/United States
- 5T32CA009161/CA/NCI NIH HHS/United States
- R56 AI070310/AI/NIAID NIH HHS/United States
- P30CA016087/CA/NCI NIH HHS/United States (VSports手机版)
"V体育安卓版" LinkOut - more resources
"V体育2025版" Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

