Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis
- PMID: 19887610
- PMCID: PMC2783591
- DOI: 10.1158/0008-5472.CAN-09-1568
Pattern of antioxidant and DNA repair gene expression in normal airway epithelium associated with lung cancer diagnosis
Abstract
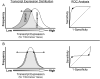
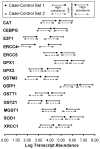
In previous studies, we reported that key antioxidant and DNA repair genes are regulated differently in normal bronchial epithelial cells of lung cancer cases compared with non-lung cancer controls. In an effort to develop a biomarker for lung cancer risk, we evaluated the transcript expressions of 14 antioxidant, DNA repair, and transcription factor genes in normal bronchial epithelial cells (HUGO names CAT, CEBPG, E2F1, ERCC4, ERCC5, GPX1, GPX3, GSTM3, GSTP1, GSTT1, GSTZ1, MGST1, SOD1, and XRCC1). A test comprising these 14 genes accurately identified the lung cancer cases in two case-control studies. The receiver operating characteristic-area under the curve was 0. 82 (95% confidence intervals, 0. 68-0. 91) for the first case-control set (25 lung cancer cases and 24 controls), and 0. 87 (95% confidence intervals, 0. 73-0. 96) for the second set (18 cases and 22 controls). For each gene included in the test, the key difference between cases and controls was altered distribution of transcript expression among cancer cases compared with controls, with more lung cancer cases expressing at both extremes among all genes (Kolmorogov-Smirnov test, D = 0. 0795; P = 0. 041). A novel statistical approach was used to identify the lower and upper boundaries of transcript expression that optimally classifies cases and controls for each gene VSports手机版. Based on the data presented here, there is an increased prevalence of lung cancer diagnosis among individuals that express a threshold number of key antioxidant, DNA repair, and transcription factor genes at either very high or very low levels in the normal airway epithelium. .
"V体育2025版" Conflict of interest statement
Conflict of Interest Statement: James C. Willey has significant equity interest in Gene Express, Inc V体育安卓版. , which produces and markets StaRT-PCRTM reagents used in this study.
V体育平台登录 - Figures


"VSports手机版" References
-
- Alberg AJ, Samet JM. Epidemiology of lung cancer. Chest. 2003;123:21S–49S. - PubMed (VSports)
-
- Gloeckler Ries LA, Reichman ME, Lewis DR, Hankey BF, Edwards BK. Cancer survival and incidence from the Surveillance, Epidemiology, and End Results (SEER) program. Oncologist. 2003;8:541–52. - PubMed
-
- Shields PG. Molecular epidemiology of lung cancer. Ann Oncol. 1999;10 (Suppl 5):S7–11. - PubMed (V体育ios版)
-
- Ganti AK, Mulshine JL. Lung cancer screening. Oncologist. 2006;11:481–7. - PubMed
Publication types
MeSH terms
- "V体育官网入口" Actions
- "VSports注册入口" Actions
- "VSports" Actions
- V体育2025版 - Actions
- Actions (VSports最新版本)
- VSports在线直播 - Actions
- "VSports注册入口" Actions
Substances
- V体育ios版 - Actions
- V体育ios版 - Actions
Grants and funding
LinkOut - more resources (V体育2025版)
"VSports app下载" Full Text Sources
Other Literature Sources
Medical
"V体育安卓版" Research Materials
Miscellaneous

