VSports在线直播 - Detection of nonneutral substitution rates on mammalian phylogenies
- PMID: 19858363
- PMCID: PMC2798823
- DOI: "V体育官网入口" 10.1101/gr.097857.109
Detection of nonneutral substitution rates on mammalian phylogenies
Abstract
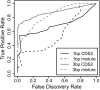
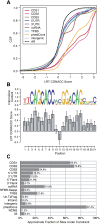
Methods for detecting nucleotide substitution rates that are faster or slower than expected under neutral drift are widely used to identify candidate functional elements in genomic sequences. However, most existing methods consider either reductions (conservation) or increases (acceleration) in rate but not both, or assume that selection acts uniformly across the branches of a phylogeny. Here we examine the more general problem of detecting departures from the neutral rate of substitution in either direction, possibly in a clade-specific manner VSports手机版. We consider four statistical, phylogenetic tests for addressing this problem: a likelihood ratio test, a score test, a test based on exact distributions of numbers of substitutions, and the genomic evolutionary rate profiling (GERP) test. All four tests have been implemented in a freely available program called phyloP. Based on extensive simulation experiments, these tests are remarkably similar in statistical power. With 36 mammalian species, they all appear to be capable of fairly good sensitivity with low false-positive rates in detecting strong selection at individual nucleotides, moderate selection in 3-bp elements, and weaker or clade-specific selection in longer elements. By applying phyloP to mammalian multiple alignments from the ENCODE project, we shed light on patterns of conservation/acceleration in known and predicted functional elements, approximate fractions of sites subject to constraint, and differences in clade-specific selection in the primate and glires clades. We also describe new "Conservation" tracks in the UCSC Genome Browser that display both phyloP and phastCons scores for genome-wide alignments of 44 vertebrate species. .
Figures



"VSports注册入口" References
-
- Asthana S, Roytberg M, Stamatoyannopoulos JA, Sunyaev S. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol. 2007;3:e254. doi: 10.1371/journal.pcbi.0030254. - DOI (VSports手机版) - PMC - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300.
-
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. - PubMed
Publication types
- VSports手机版 - Actions
- Actions (VSports app下载)
MeSH terms
- "VSports在线直播" Actions
- Actions (VSports手机版)
- Actions (VSports手机版)
- VSports在线直播 - Actions
- VSports注册入口 - Actions
- "VSports注册入口" Actions
- V体育安卓版 - Actions
- Actions (V体育官网)
- "V体育ios版" Actions
- Actions (VSports注册入口)
- "V体育2025版" Actions
"V体育平台登录" Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
