Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition
- PMID: 19776130
- PMCID: PMC2786736
- DOI: 10.1128/JVI.01491-09
Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition
Abstract
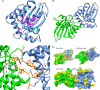
Retroelements are important evolutionary forces but can be deleterious if left uncontrolled. Members of the human APOBEC3 family of cytidine deaminases can inhibit a wide range of endogenous, as well as exogenous, retroelements. These enzymes are structurally organized in one or two domains comprising a zinc-coordinating motif VSports手机版. APOBEC3G contains two such domains, only the C terminal of which is endowed with editing activity, while its N-terminal counterpart binds RNA, promotes homo-oligomerization, and is necessary for packaging into human immunodeficiency virus type 1 (HIV-1) virions. Here, we performed a large-scale mutagenesis-based analysis of the APOBEC3G N terminus, testing mutants for (i) inhibition of vif-defective HIV-1 infection and Alu retrotransposition, (ii) RNA binding, and (iii) oligomerization. Furthermore, in the absence of structural information on this domain, we used homology modeling to examine the positions of functionally important residues and of residues found to be under positive selection by phylogenetic analyses of primate APOBEC3G genes. Our results reveal the importance of a predicted RNA binding dimerization interface both for packaging into HIV-1 virions and inhibition of both HIV-1 infection and Alu transposition. We further found that the HIV-1-blocking activity of APOBEC3G N-terminal mutants defective for packaging can be almost entirely rescued if their virion incorporation is forced by fusion with Vpr, indicating that the corresponding region of APOBEC3G plays little role in other aspects of its action against this pathogen. Interestingly, residues forming the APOBEC3G dimer interface are highly conserved, contrasting with the rapid evolution of two neighboring surface-exposed amino acid patches, one targeted by the Vif protein of primate lentiviruses and the other of yet-undefined function. .
Figures

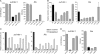


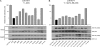

References
-
- Ao, Z., Z. Yu, L. Wang, Y. Zheng, and X. Yao. 2008. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS ONE 3:e1995. - PMC (VSports) - PubMed
-
- Berman, H., K. Henrick, H. Nakamura, and J. L. Markley. 2007. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 35:D301-D303. - "V体育平台登录" PMC - PubMed
-
- Bogerd, H. P., and B. R. Cullen. 2008. Single-stranded RNA facilitates nucleocapsid: APOBEC3G complex formation. RNA 14:1228-1236. - "VSports在线直播" PMC - PubMed
-
- Bordo, D., and P. Argos. 1991. Suggestions for “safe” residue substitutions in site-directed mutagenesis. J. Mol. Biol. 217:721-729. - PubMed
Publication types
MeSH terms
- V体育ios版 - Actions
- Actions (V体育官网)
- "VSports在线直播" Actions
- VSports - Actions
- "V体育官网入口" Actions
- Actions (V体育官网)
- "V体育安卓版" Actions
- VSports注册入口 - Actions
- Actions (V体育平台登录)
Substances
- V体育ios版 - Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

