A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase
- PMID: 19561087
- PMCID: PMC2755697
- DOI: 10.1074/jbc.M109.025536
"VSports最新版本" A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase
Abstract
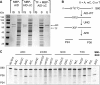
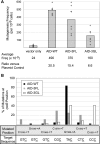
Enzymes of the AID/APOBEC family, characterized by the targeted deamination of cytosine to generate uracil within DNA, mediate numerous critical immune responses. One family member, activation-induced cytidine deaminase (AID), selectively introduces uracil into antibody variable and switch regions, promoting antibody diversity through somatic hypermutation or class switching. Other family members, including APOBEC3F and APOBEC3G, play an important role in retroviral defense by acting on viral reverse transcripts. These enzymes are distinguished from one another by targeting cytosine within different DNA sequence contexts; however, the reason for these differences is not known. Here, we report the identification of a recognition loop of 9-11 amino acids that contributes significantly to the distinct sequence motifs of individual family members. When this recognition loop is grafted from the donor APOBEC3F or 3G proteins into the acceptor scaffold of AID, the mutational signature of AID changes toward that of the donor proteins. These loop-graft mutants of AID provide useful tools for dissecting the biological impact of DNA sequence preferences upon generation of antibody diversity, and the results have implications for the evolution and specialization of the AID/APOBEC family VSports手机版. .
Figures
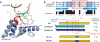

References
-
- Muramatsu M., Nagaoka H., Shinkura R., Begum N. A., Honjo T. (2007) Adv. Immunol. 94,1–36 - PubMed
-
- Conticello S. G., Langlois M. A., Yang Z., Neuberger M. S. (2007) Adv. Immunol. 94,37–73 - VSports最新版本 - PubMed
-
- Rosenberg B. R., Papavasiliou F. N. (2007) Adv. Immunol. 94,215–244 - PubMed
-
- Muramatsu M., Sankaranand V. S., Anant S., Sugai M., Kinoshita K., Davidson N. O., Honjo T. (1999) J. Biol. Chem. 274,18470–18476 - PubMed
-
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., Tezcan I., Ersoy F., Kayserili H., Ugazio A. G., Brousse N., Muramatsu M., Notarangelo L. D., Kinoshita K., Honjo T., Fischer A., Durandy A. (2000) Cell 102,565–575 - "VSports最新版本" PubMed
Publication types (VSports注册入口)
VSports最新版本 - MeSH terms
- "VSports注册入口" Actions
- Actions (VSports app下载)
- Actions (VSports注册入口)
- Actions (V体育官网入口)
- V体育ios版 - Actions
- "V体育官网" Actions
- V体育安卓版 - Actions
- Actions (VSports注册入口)
VSports注册入口 - Substances
- "VSports手机版" Actions
VSports最新版本 - Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

