V体育安卓版 - Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths
- PMID: 19165319
- PMCID: PMC2617782
- DOI: "V体育ios版" 10.1371/journal.pgen.1000344
Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths (VSports最新版本)
Abstract
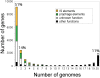
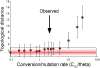
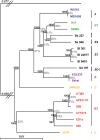
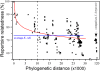
The Escherichia coli species represents one of the best-studied model organisms, but also encompasses a variety of commensal and pathogenic strains that diversify by high rates of genetic change. We uniformly (re-) annotated the genomes of 20 commensal and pathogenic E. coli strains and one strain of E. fergusonii (the closest E. coli related species), including seven that we sequenced to completion. Within the approximately 18,000 families of orthologous genes, we found approximately 2,000 common to all strains. Although recombination rates are much higher than mutation rates, we show, both theoretically and using phylogenetic inference, that this does not obscure the phylogenetic signal, which places the B2 phylogenetic group and one group D strain at the basal position. Based on this phylogeny, we inferred past evolutionary events of gain and loss of genes, identifying functional classes under opposite selection pressures. We found an important adaptive role for metabolism diversification within group B2 and Shigella strains, but identified few or no extraintestinal virulence-specific genes, which could render difficult the development of a vaccine against extraintestinal infections. Genome flux in E VSports手机版. coli is confined to a small number of conserved positions in the chromosome, which most often are not associated with integrases or tRNA genes. Core genes flanking some of these regions show higher rates of recombination, suggesting that a gene, once acquired by a strain, spreads within the species by homologous recombination at the flanking genes. Finally, the genome's long-scale structure of recombination indicates lower recombination rates, but not higher mutation rates, at the terminus of replication. The ensuing effect of background selection and biased gene conversion may thus explain why this region is A+T-rich and shows high sequence divergence but low sequence polymorphism. Overall, despite a very high gene flow, genes co-exist in an organised genome. .
"VSports在线直播" Conflict of interest statement
The authors have declared that no competing interests exist.
"VSports在线直播" Figures




V体育2025版 - References
-
- Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F, et al., editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, DC: American Society for Microbiology; 2004. [Online.] http://www.ecosal.org.
-
- Hobman JL, Penn CW, Pallen MJ. Laboratory strains of Escherichia coli: model citizens or deceitful delinquents growing old disgracefully? Molecular Microbiol. 2007;64:881–885. - PubMed
-
- Savageau MA. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat. 1983;122:732–744.
-
- Donnenberg MS. Escherichia coli. Virulence mechanisms of a versatile pathogen. Baltimore: Academic press, Elsevier Science; 2002.
-
- Rolland K, Lambert-Zeschovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli strains are derived from distinct ancestral strains of E. coli. Microbiology. 1998;144:2667–2672. - PubMed
Publication types
- Actions (V体育官网入口)
"VSports app下载" MeSH terms
- "VSports最新版本" Actions
- Actions (VSports最新版本)
- "VSports在线直播" Actions
Substances
- Actions (V体育2025版)
"V体育官网" LinkOut - more resources
Full Text Sources
Other Literature Sources
VSports app下载 - Molecular Biology Databases
Research Materials
Miscellaneous

