Small RNAs in transcriptional gene silencing and genome defence
- PMID: 19158787
- PMCID: "V体育官网入口" PMC3246369
- DOI: 10.1038/nature07756
Small RNAs in transcriptional gene silencing and genome defence
Abstract (VSports最新版本)
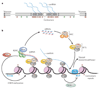
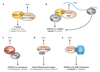
Small RNA molecules of about 20-30 nucleotides have emerged as powerful regulators of gene expression and genome stability. Studies in fission yeast and multicellular organisms suggest that effector complexes, directed by small RNAs, target nascent chromatin-bound non-coding RNAs and recruit chromatin-modifying complexes VSports手机版. Interactions between small RNAs and nascent non-coding transcripts thus reveal a new mechanism for targeting chromatin-modifying complexes to specific chromosome regions and suggest possibilities for how the resultant chromatin states may be inherited during the process of chromosome duplication. .
Figures

References
-
- Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. - PubMed
-
- Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nature Struct. Mol. Biol. 2007;14:1041–1048. - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - "VSports在线直播" PubMed
-
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. - PubMed
"VSports最新版本" Publication types
- "VSports" Actions
- Actions (V体育安卓版)
- "VSports app下载" Actions
MeSH terms
- V体育安卓版 - Actions
- Actions (V体育2025版)
- "V体育安卓版" Actions
- Actions (V体育ios版)
- VSports最新版本 - Actions
Substances
- Actions (VSports注册入口)
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

