Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing
- PMID: 19023400
- PMCID: PMC2577301
- DOI: 10.1371/journal.pgen.1000255
Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing
Erratum in
- PLoS Genet. 2008 Dec;4(12). doi: 10.1371/annotation/3d8a6578-ce56-45aa-bc71-05078355b851. Welch, David Mark [corrected to Mark Welch, David]
V体育安卓版 - Abstract
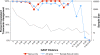
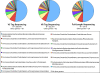
Massively parallel pyrosequencing of hypervariable regions from small subunit ribosomal RNA (SSU rRNA) genes can sample a microbial community two or three orders of magnitude more deeply per dollar and per hour than capillary sequencing of full-length SSU rRNA. As with full-length rRNA surveys, each sequence read is a tag surrogate for a single microbe. However, rather than assigning taxonomy by creating gene trees de novo that include all experimental sequences and certain reference taxa, we compare the hypervariable region tags to an extensive database of rRNA sequences and assign taxonomy based on the best match in a Global Alignment for Sequence Taxonomy (GAST) process. The resulting taxonomic census provides information on both composition and diversity of the microbial community. To determine the effectiveness of using only hypervariable region tags for assessing microbial community membership, we compared the taxonomy assigned to the V3 and V6 hypervariable regions with the taxonomy assigned to full-length SSU rRNA sequences isolated from both the human gut and a deep-sea hydrothermal vent. The hypervariable region tags and full-length rRNA sequences provided equivalent taxonomy and measures of relative abundance of microbial communities, even for tags up to 15% divergent from their nearest reference match. The greater sampling depth per dollar afforded by massively parallel pyrosequencing reveals many more members of the "rare biosphere" than does capillary sequencing of the full-length gene. In addition, tag sequencing eliminates cloning bias and the sequences are short enough to be completely sequenced in a single read, maximizing the number of organisms sampled in a run while minimizing chimera formation. This technique allows the cost-effective exploration of changes in microbial community structure, including the rare biosphere, over space and time and can be applied immediately to initiatives, such as the Human Microbiome Project VSports手机版. .
Conflict of interest statement (VSports手机版)
The authors have declared that no competing interests exist.
"V体育安卓版" Figures


References
-
- Atlas RM, Bartha R. Microbial Ecology: Fundamentals and Applications. Redwood City: The Benjamin/Cummings Publishing Company, Inc; 1993.
-
- Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, et al. Metagenomic and Small-Subunit rRNA Analyses Reveal the Genetic Diversity of Bacteria, Archaea, Fungi, and Viruses in Soil. Appl Environ Microbiol. 2007;73:7059–7066. - "V体育官网入口" PMC - PubMed
-
- Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. - "VSports在线直播" PMC - PubMed
Publication types
- "V体育官网" Actions
- V体育ios版 - Actions
- Actions (V体育平台登录)
MeSH terms (VSports app下载)
- "VSports注册入口" Actions
- Actions (VSports在线直播)
- V体育官网入口 - Actions
VSports在线直播 - Substances
Grants and funding
LinkOut - more resources (VSports最新版本)
Full Text Sources
Other Literature Sources
Research Materials

