Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53
- PMID: 19020049
- PMCID: "V体育2025版" PMC6671724
- DOI: 10.1523/JNEUROSCI.4048-08.2008
Neuronal death resulting from targeted disruption of the Snf2 protein ATRX is mediated by p53
Abstract
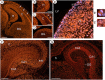
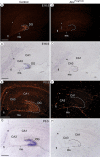
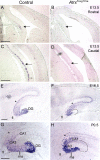
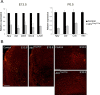
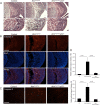
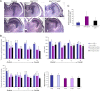
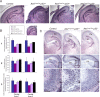
ATRX, a chromatin remodeling protein of the Snf2 family, participates in diverse cellular functions including regulation of gene expression and chromosome alignment during mitosis and meiosis. Mutations in the human gene cause alpha thalassemia mental retardation, X-linked (ATR-X) syndrome, a rare disorder characterized by severe cognitive deficits, microcephaly and epileptic seizures VSports手机版. Conditional inactivation of the Atrx gene in the mouse forebrain leads to neonatal lethality and defective neurogenesis manifested by increased cell death and reduced cellularity in the developing neocortex and hippocampus. Here, we show that Atrx-null forebrains do not generate dentate granule cells due to a reduction in precursor cell number and abnormal migration of differentiating granule cells. In addition, fewer GABA-producing interneurons are generated that migrate from the ventral telencephalon to the cortex and hippocampus. Staining for cleaved caspase 3 demonstrated increased apoptosis in both the hippocampal hem and basal telencephalon concurrent with p53 pathway activation. Elimination of the tumor suppressor protein p53 in double knock-out mice rescued cell death in the embryonic telencephalon but only partially ameliorated the Atrx-null phenotypes at birth. Together, these findings show that ATRX deficiency leads to p53-dependent neuronal apoptosis which is responsible for some but not all of the phenotypic consequences of ATRX deficiency in the forebrain. .
"V体育官网入口" Figures

References
-
- Abidi F, Schwartz CE, Carpenter NJ, Villard L, Fontés M, Curtis M. Carpenter-Waziri syndrome results from a mutation in XNP. Am J Med Genet. 1999;85:249–251. - PubMed
-
- Altman J, Bayer SA. Migration and distribution of two populations of hippocampal granule cell precursors during the perinatal and postnatal periods. J Comp Neurol. 1990a;301:365–381. - PubMed
-
- Altman J, Bayer SA. Mosaic organization of the hippocampal neuroepithelium and the multiple germinal sources of dentate granule cells. J Comp Neurol. 1990b;301:325–342. - PubMed
-
- Bagri A, Gurney T, He X, Zou YR, Littman DR, Tessier-Lavigne M, Pleasure SJ. The chemokine SDF1 regulates migration of dentate granule cells. Development. 2002;129:4249–4260. - "V体育平台登录" PubMed
-
- Bérubé NG, Smeenk CA, Picketts DJ. Cell cycle-dependant phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Human Molecular Genetics. 2000;9:539–547. - PubMed
"VSports在线直播" Publication types
MeSH terms
- Actions (VSports app下载)
- "V体育安卓版" Actions
- Actions (VSports注册入口)
- VSports在线直播 - Actions
- "V体育ios版" Actions
- VSports手机版 - Actions
- "V体育ios版" Actions
- "V体育2025版" Actions
- Actions (V体育官网)
- Actions (VSports在线直播)
- V体育官网入口 - Actions
- "V体育安卓版" Actions
- VSports - Actions
- Actions (VSports在线直播)
- "V体育平台登录" Actions
"V体育ios版" Substances
- VSports在线直播 - Actions
- V体育ios版 - Actions
- "VSports手机版" Actions
- V体育2025版 - Actions
- "V体育平台登录" Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous (VSports最新版本)
