SCL and associated proteins distinguish active from repressive GATA transcription factor complexes
- PMID: 19011221
- PMCID: PMC2652367 (VSports app下载)
- DOI: 10.1182/blood-2008-07-169417
SCL and associated proteins distinguish active from repressive GATA transcription factor complexes
Abstract
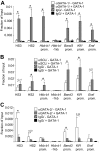
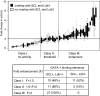
GATA-1 controls hematopoietic development by activating and repressing gene transcription, yet the in vivo mechanisms that specify these opposite activities are unknown. By examining the composition of GATA-1-associated protein complexes in a conditional erythroid rescue system as well as through the use of tiling arrays we detected the SCL/TAL1, LMO2, Ldb1, E2A complex at all positively acting GATA-1-bound elements examined VSports手机版. Similarly, the SCL complex is present at all activating GATA elements in megakaryocytes and mast cells. In striking contrast, at sites where GATA-1 functions as a repressor, the SCL complex is depleted. A DNA-binding defective form of SCL maintains association with a subset of active GATA elements indicating that GATA-1 is a key determinant for SCL recruitment. Knockdown of LMO2 selectively impairs activation but not repression by GATA-1. ETO-2, an SCL-associated protein with the potential for transcription repression, is also absent from GATA-1-repressed genes but, unlike SCL, fails to accumulate at GATA-1-activated genes. Together, these studies identify the SCL complex as a critical and consistent determinant of positive GATA-1 activity in multiple GATA-1-regulated hematopoietic cell lineages. .
Figures

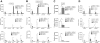



VSports - References
-
- Nishiyama C, Ito T, Nishiyama M, et al. GATA-1 is required for expression of FcεRI on mast cells: analysis of mast cells derived from GATA-1 knockdown mouse bone marrow. Int Immunol. 2005;17:847–856. - PubMed (V体育官网入口)
Publication types
- V体育2025版 - Actions
MeSH terms
- "VSports在线直播" Actions
- "V体育2025版" Actions
- V体育官网 - Actions
- Actions (V体育官网入口)
- "V体育官网" Actions
- "VSports在线直播" Actions
- Actions (VSports最新版本)
- "V体育官网入口" Actions
- V体育平台登录 - Actions
Substances
- VSports注册入口 - Actions
- VSports app下载 - Actions
- "V体育安卓版" Actions
- "V体育平台登录" Actions
- "V体育2025版" Actions
- Actions (V体育平台登录)
- VSports在线直播 - Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
VSports注册入口 - Research Materials
"VSports" Miscellaneous

