The diploid genome sequence of an Asian individual
- PMID: 18987735
- PMCID: PMC2716080
- DOI: 10.1038/nature07484
The diploid genome sequence of an Asian individual (VSports)
Abstract
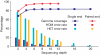
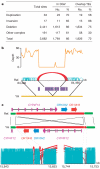
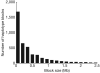
Here we present the first diploid genome sequence of an Asian individual. The genome was sequenced to 36-fold average coverage using massively parallel sequencing technology. We aligned the short reads onto the NCBI human reference genome to 99. 97% coverage, and guided by the reference genome, we used uniquely mapped reads to assemble a high-quality consensus sequence for 92% of the Asian individual's genome. We identified approximately 3 million single-nucleotide polymorphisms (SNPs) inside this region, of which 13. 6% were not in the dbSNP database. Genotyping analysis showed that SNP identification had high accuracy and consistency, indicating the high sequence quality of this assembly. We also carried out heterozygote phasing and haplotype prediction against HapMap CHB and JPT haplotypes (Chinese and Japanese, respectively), sequence comparison with the two available individual genomes (J. D. Watson and J. C VSports手机版. Venter), and structural variation identification. These variations were considered for their potential biological impact. Our sequence data and analyses demonstrate the potential usefulness of next-generation sequencing technologies for personal genomics. .
Figures

Comment in (VSports注册入口)
-
Human genetics: Individual genomes diversify.Nature. 2008 Nov 6;456(7218):49-51. doi: 10.1038/456049a. Nature. 2008. PMID: 18987731 No abstract available.
"V体育安卓版" References
-
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed (V体育平台登录)
-
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
"V体育ios版" Publication types
- "VSports在线直播" Actions
MeSH terms
- VSports手机版 - Actions
- "V体育官网入口" Actions
- "VSports" Actions
- Actions (VSports app下载)
- VSports在线直播 - Actions
- "V体育安卓版" Actions
Grants and funding
"VSports注册入口" LinkOut - more resources
VSports注册入口 - Full Text Sources
Other Literature Sources

