Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
- PMID: 18845756
- PMCID: PMC3272704
- DOI: 10.1126/science.1162403
Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain
Abstract
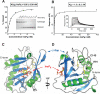
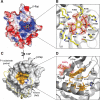
Vibrio cholerae RTX (repeats in toxin) is an actin-disrupting toxin that is autoprocessed by an internal cysteine protease domain (CPD). The RTX CPD is efficiently activated by the eukaryote-specific small molecule inositol hexakisphosphate (InsP6), and we present the 2. 1 angstrom structure of the RTX CPD in complex with InsP6. InsP6 binds to a conserved basic cleft that is distant from the protease active site VSports手机版. Biochemical and kinetic analyses of CPD mutants indicate that InsP6 binding induces an allosteric switch that leads to the autoprocessing and intracellular release of toxin-effector domains. .
V体育2025版 - Figures

V体育安卓版 - References
-
- Reineke J, et al. Nature. 2007;446:415. - "VSports" PubMed
-
- Cordero CL, Sozhamannan S, Satchell KJ. J. Clin. Microbiol. 2007;45:2289. - PMC (V体育平台登录) - PubMed
Publication types
"V体育平台登录" MeSH terms
- Actions (VSports app下载)
- Actions (V体育ios版)
- "VSports注册入口" Actions
- "V体育官网入口" Actions
- VSports手机版 - Actions
- "VSports app下载" Actions
- Actions (V体育官网)
- VSports最新版本 - Actions
- "V体育安卓版" Actions
- VSports手机版 - Actions
- "VSports注册入口" Actions
- "VSports最新版本" Actions
- "V体育官网" Actions
- Actions (VSports手机版)
- Actions (VSports手机版)
- "V体育官网入口" Actions
Substances
- "VSports最新版本" Actions
- V体育官网入口 - Actions
V体育官网入口 - Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
V体育安卓版 - Molecular Biology Databases

