"V体育2025版" Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome
- PMID: 18698407
- PMCID: PMC2492807 (V体育平台登录)
- DOI: 10.1371/journal.pone.0002945
Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome
Abstract
Background: The complex microbiome of the ceca of chickens plays an important role in nutrient utilization, growth and well-being of these animals. Since we have a very limited understanding of the capabilities of most species present in the cecum, we investigated the role of the microbiome by comparative analyses of both the microbial community structure and functional gene content using random sample pyrosequencing. The overall goal of this study was to characterize the chicken cecal microbiome using a pathogen-free chicken and one that had been challenged with Campylobacter jejuni VSports手机版. .
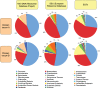
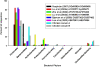
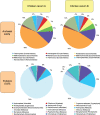
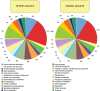
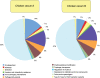
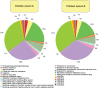
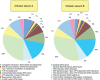
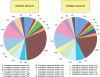
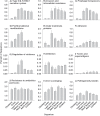
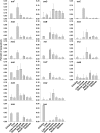
Methodology/principal findings: Comparative metagenomic pyrosequencing was used to generate 55,364,266 bases of random sampled pyrosequence data from two chicken cecal samples. SSU rDNA gene tags and environmental gene tags (EGTs) were identified using SEED subsystems-based annotations. The distribution of phylotypes and EGTs detected within each cecal sample were primarily from the Firmicutes, Bacteroidetes and Proteobacteria, consistent with previous SSU rDNA libraries of the chicken cecum. Carbohydrate metabolism and virulence genes are major components of the EGT content of both of these microbiomes. A comparison of the twelve major pathways in the SEED Virulence Subsystem (metavirulome) represented in the chicken cecum, mouse cecum and human fecal microbiomes showed that the metavirulomes differed between these microbiomes and the metavirulomes clustered by host environment. The chicken cecum microbiomes had the broadest range of EGTs within the SEED Conjugative Transposon Subsystem, however the mouse cecum microbiomes showed a greater abundance of EGTs in this subsystem V体育安卓版. Gene assemblies (32 contigs) from one microbiome sample were predominately from the Bacteroidetes, and seven of these showed sequence similarity to transposases, whereas the remaining sequences were most similar to those from catabolic gene families. .
Conclusion/significance: This analysis has demonstrated that mobile DNA elements are a major functional component of cecal microbiomes, thus contributing to horizontal gene transfer and functional microbiome evolution. Moreover, the metavirulomes of these microbiomes appear to associate by host environment. These data have implications for defining core and variable microbiome content in a host species V体育ios版. Furthermore, this suggests that the evolution of host specific metavirulomes is a contributing factor in disease resistance to zoonotic pathogens. .
Conflict of interest statement
Figures

"V体育2025版" References
-
- Hyman ED. A New Method of Sequencing DNA. Analytical Biochemistry. 1988;174:423–436. - PubMed
-
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. - "V体育平台登录" PubMed
-
- Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Analytical Biochemistry. 1996;242:84–89. - PubMed
Publication types (V体育2025版)
MeSH terms
- Actions (VSports手机版)
- "V体育安卓版" Actions
- "V体育安卓版" Actions
- "V体育平台登录" Actions
- "V体育2025版" Actions
- Actions (VSports)
- VSports - Actions
- Actions (VSports在线直播)
- Actions (V体育平台登录)
- Actions (VSports最新版本)
- "V体育平台登录" Actions
- "V体育安卓版" Actions
Substances
- Actions (VSports手机版)

