VSports - Group contribution method for thermodynamic analysis of complex metabolic networks
- PMID: 18645197
- PMCID: PMC2479599
- DOI: 10.1529/biophysj.107.124784
Group contribution method for thermodynamic analysis of complex metabolic networks
"V体育平台登录" Abstract
A new, to our knowledge, group contribution method based on the group contribution method of Mavrovouniotis is introduced for estimating the standard Gibbs free energy of formation (Delta(f)G'(o)) and reaction (Delta(r)G'(o)) in biochemical systems. Gibbs free energy contribution values were estimated for 74 distinct molecular substructures and 11 interaction factors using multiple linear regression against a training set of 645 reactions and 224 compounds. The standard error for the fitted values was 1. 90 kcal/mol. Cross-validation analysis was utilized to determine the accuracy of the methodology in estimating Delta(r)G'(o) and Delta(f)G'(o) for reactions and compounds not included in the training set, and based on the results of the cross-validation, the standard error involved in these estimations is 2. 22 kcal/mol. This group contribution method is demonstrated to be capable of estimating Delta(r)G'(o) and Delta(f)G'(o) for the majority of the biochemical compounds and reactions found in the iJR904 and iAF1260 genome-scale metabolic models of Escherichia coli and in the Kyoto Encyclopedia of Genes and Genomes and University of Minnesota Biocatalysis and Biodegradation Database. A web-based implementation of this new group contribution method is available free at http://sparta VSports手机版. chem-eng. northwestern. edu/cgi-bin/GCM/WebGCM. cgi. .
Figures
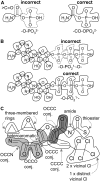

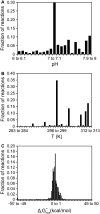

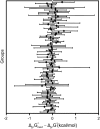
References
-
- Price, N. D., J. A. Papin, C. H. Schilling, and B. O. Palsson. 2003. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 21:162–169. - "V体育ios版" PubMed
-
- Feist, A. M., C. S. Henry, J. L. Reed, M. Krummenacker, A. R. Joyce, P. D. Karp, L. J. Broadbelt, V. Hatzimanikatis, and B. Ø. Palsson. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1261 ORFs and thermodynamic information. Mol. Syst. Biol. 3:1–18. - PMC - PubMed
Publication types
- Actions (V体育2025版)
- "V体育官网" Actions
MeSH terms
- VSports在线直播 - Actions
- VSports最新版本 - Actions
- VSports最新版本 - Actions
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

