Molecular signatures of metastasis in head and neck cancer
- PMID: 18642293
- PMCID: PMC4136479
- DOI: 10.1002/hed.20871
Molecular signatures of metastasis in head and neck cancer
Abstract
Background: Metastases are the primary cause of cancer treatment failure and death, yet metastatic mechanisms remain incompletely understood. VSports手机版.
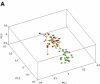
Methods: We studied the molecular basis of head and neck cancer metastasis by transcriptionally profiling 70 samples from 27 patients-matching normal adjacent tissue, primary tumor, and cervical lymph node metastases. V体育安卓版.
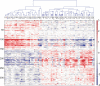
Results: We identified tumor-associated expression signatures common to both primary tumors and metastases. Use of matching metastases revealed an additional 46 dysregulated genes associated solely with head and neck cancer metastasis. However, despite being metastasis-specific in our sample set, these 46 genes are concordant with genes previously discovered in primary tumors that metastasized. V体育ios版.
Conclusions: Although our data and related studies show that most of the metastatic potential appears to be inherent to the primary tumor, they are also consistent with the notion that a limited number of additional clonal changes are necessary to yield the final metastatic cell(s), albeit in a variable temporal order. VSports最新版本.
Copyright (c) 2008 Wiley Periodicals, Inc V体育平台登录. Head Neck 2008. .
Figures


References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. - "VSports注册入口" PubMed
-
- Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–297. - VSports最新版本 - PubMed
-
- Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. - PubMed (VSports在线直播)
-
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. - PubMed
Publication types
- Actions (V体育ios版)
- V体育2025版 - Actions
VSports app下载 - MeSH terms
- "V体育安卓版" Actions
- "V体育2025版" Actions
- VSports app下载 - Actions
- "V体育官网入口" Actions
Grants and funding
LinkOut - more resources (VSports app下载)
VSports在线直播 - Full Text Sources
Other Literature Sources
Medical

