V体育2025版 - Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2
- PMID: 18641123
- PMCID: PMC2492495 (VSports最新版本)
- DOI: 10.1073/pnas.0804453105 (VSports手机版)
Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2
VSports在线直播 - Abstract
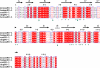
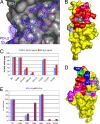
Programmed death-1 (PD-1) is a member of the CD28/B7 superfamily that delivers negative signals upon interaction with its two ligands, PD-L1 or PD-L2. The high-resolution crystal structure of the complex formed by the complete ectodomains of murine PD-1 and PD-L2 revealed a 1:1 receptor:ligand stoichiometry and displayed a binding interface and overall molecular organization distinct from that observed in the CTLA-4/B7 inhibitory complexes. Furthermore, our structure also provides insights into the association between PD-1 and PD-L1 and highlights differences in the interfaces formed by the two PD-1 ligands (PD-Ls) Mutagenesis studies confirmed the details of the proposed PD-1/PD-L binding interfaces and allowed for the design of a mutant PD-1 receptor with enhanced affinity. These studies define spatial and organizational constraints that control the localization and signaling of PD-1/PD-L complexes within the immunological synapse and provide a basis for manipulating the PD-1 pathways for immunotherapy. VSports手机版.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


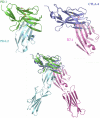

VSports app下载 - Comment in
-
"VSports在线直播" Structures of PD-1 with its ligands: sideways and dancing cheek to cheek.Proc Natl Acad Sci U S A. 2008 Jul 29;105(30):10275-6. doi: 10.1073/pnas.0805459105. Epub 2008 Jul 23. Proc Natl Acad Sci U S A. 2008. PMID: 18650389 Free PMC article. No abstract available.
References
-
- Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. - PubMed
-
- Okazaki T, Honjo T. PD-1 and PD-1 ligands: From discovery to clinical application. Int Immunol. 2007;19:813–824. - V体育2025版 - PubMed
-
- Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. - PubMed
Publication types (V体育ios版)
- Actions (VSports app下载)
"VSports app下载" MeSH terms
- VSports注册入口 - Actions
- Actions (V体育ios版)
- Actions (VSports最新版本)
- "V体育平台登录" Actions
- VSports - Actions
- Actions (V体育2025版)
"V体育官网" Substances
- Actions (VSports)
- V体育平台登录 - Actions
- "V体育安卓版" Actions
Associated data
- Actions (V体育ios版)
- VSports - Actions
Grants and funding
VSports手机版 - LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

