"V体育官网" Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA
- PMID: 18603530
- PMCID: "V体育官网入口" PMC3259822
- DOI: 10.1074/jbc.M803776200
Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA
V体育官网入口 - Abstract
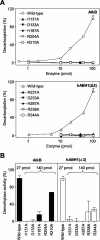
The Escherichia coli AlkB protein and human homologs hABH2 and hABH3 are 2-oxoglutarate (2OG)/Fe(II)-dependent DNA/RNA demethylases that repair 1-methyladenine and 3-methylcytosine residues. Surprisingly, hABH1, which displays the strongest homology to AlkB, failed to show repair activity in two independent studies. Here, we show that hABH1 is a mitochondrial protein, as demonstrated using fluorescent fusion protein expression, immunocytochemistry, and Western blot analysis. A fraction is apparently nuclear and this fraction increases strongly if the fluorescent tag is placed at the N-terminal end of the protein, thus interfering with mitochondrial targeting. Molecular modeling of hABH1 based upon the sequence and known structures of AlkB and hABH3 suggested an active site almost identical to these enzymes. hABH1 decarboxylates 2OG in the absence of a prime substrate, and the activity is stimulated by methylated nucleotides. Employing three different methods we demonstrate that hABH1 demethylates 3-methylcytosine in single-stranded DNA and RNA in vitro. Site-specific mutagenesis confirmed that the putative Fe(II) and 2OG binding residues are essential for activity VSports手机版. In conclusion, hABH1 is a functional mitochondrial AlkB homolog that repairs 3-methylcytosine in single-stranded DNA and RNA. .
Figures





"VSports手机版" References
-
- Drabløs, F., Feyzi, E., Aas, P. A., Vaagbø, C. B., Kavli, B., Bratlie, M. S., Peña-Diaz, J., Otterlei, M., Slupphaug, G., and Krokan, H. E. (2004) DNA Repair 31389 -1407 - PubMed
-
- Sedgwick, B. (2004) Nat. Rev. Mol. Cell Biol. 5148 -157 - PubMed (VSports注册入口)
-
- Sedgwick, B., and Lindahl, T. (2002) Oncogene 218886 -8894 - PubMed
-
- Falnes, P. O., Johansen, R. F., and Seeberg, E. (2002) Nature 419178 -182 - VSports app下载 - PubMed
VSports手机版 - Publication types
MeSH terms
- VSports app下载 - Actions
- Actions (V体育ios版)
- Actions (V体育平台登录)
- Actions (V体育ios版)
- Actions (VSports在线直播)
- VSports手机版 - Actions
- Actions (V体育官网入口)
- Actions (V体育官网)
- "V体育官网入口" Actions
VSports在线直播 - Substances
- Actions (V体育官网入口)
- Actions (VSports在线直播)
- "VSports app下载" Actions
- VSports最新版本 - Actions
- "V体育ios版" Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

