Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen
- PMID: 18519643
- PMCID: PMC2418588
- DOI: 10.1101/gad.1652308
Identification of novel genes involved in light-dependent CRY degradation through a genome-wide RNAi screen
Abstract
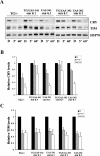
Circadian clocks regulate many different physiological processes and synchronize these to environmental light:dark cycles. In Drosophila, light is transmitted to the clock by a circadian blue light photoreceptor CRYPTOCHROME (CRY). In response to light, CRY promotes the degradation of the circadian clock protein TIMELESS (TIM) and then is itself degraded. To identify novel genes involved in circadian entrainment, we performed an unbiased genome-wide screen in Drosophila cells using a sensitive and quantitative assay that measures light-induced degradation of CRY. We systematically knocked down the expression of approximately 21,000 genes and identified those that regulate CRY stability. These genes include ubiquitin ligases, signal transduction molecules, and redox molecules. Many of the genes identified in the screen are specific for CRY degradation and do not affect degradation of the TIM protein in response to light, suggesting that, for the most part, these two pathways are distinct. We further validated the effect of three candidate genes on CRY stability in vivo by assaying flies mutant for each of these genes. This work identifies a novel regulatory network involved in light-dependent CRY degradation and demonstrates the power of a genome-wide RNAi approach for understanding circadian biology VSports手机版. .
Figures




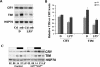
"VSports app下载" References
-
- Adams J.D., Klaidman L.K., Ribeiro P. Tyrosine hydroxylase: Mechanisms of oxygen radical formation. Redox Rep. 1997;3:273–279. - PubMed (V体育平台登录)
-
- Ashmore L.J., Sehgal A. A fly’s eye view of circadian entrainment. J. Biol. Rhythms. 2003;18:206–216. - PubMed
-
- Bartke T., Pohl C., Pyrowolakis G., Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell. 2004;14:801–811. - PubMed
-
- Busza A., Emery-Le M., Rosbash M., Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. - PubMed (V体育安卓版)
-
- Cashmore A.R. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. - "V体育2025版" PubMed
Publication types (V体育安卓版)
- V体育官网入口 - Actions
"V体育官网入口" MeSH terms
- Actions (V体育2025版)
- Actions (VSports app下载)
- "VSports" Actions
- V体育ios版 - Actions
- Actions (VSports app下载)
"V体育ios版" Substances
- V体育2025版 - Actions
Grants and funding
"VSports手机版" LinkOut - more resources
Full Text Sources
Molecular Biology Databases
